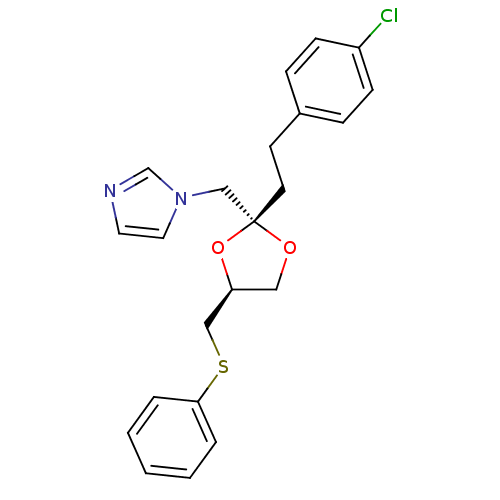
BDBM31652 imidazole-dioxolane, 3
SMILES Clc1ccc(CC[C@@]2(Cn3ccnc3)OC[C@@H](CSc3ccccc3)O2)cc1
InChI Key InChIKey=RZJAKIZOPWLXJW-RBBKRZOGSA-N
Data 4 IC50
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 4 hits for monomerid = 31652
Found 4 hits for monomerid = 31652
Affinity DataIC50: 1.03E+3nMpH: 7.4 T: 2°CAssay Description:HO activity in rat spleen (HO-1) and brain (HO-2) microsomal fractions was determined by the quantitation of CO formed from the degradation of methem...More data for this Ligand-Target Pair
Affinity DataIC50: 2.00E+3nMAssay Description:CYP3A1/3A2-catalyzed erythromycin N-demethylase activity was determined by the spectrophotometric measurement of formaldehyde. Briefly, reaction mixt...More data for this Ligand-Target Pair
Affinity DataIC50: 3.40E+4nMpH: 7.4Assay Description:HO activity in rat spleen (HO-1) and brain (HO-2) microsomal fractions was determined by the quantitation of CO formed from the degradation of methem...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:CYP2E1 hydroxylation of p-nitrophenol was determined by the spectrophotometric measurement of 4-nitrocatechol. Briefly, reaction mixture consisting o...More data for this Ligand-Target Pair