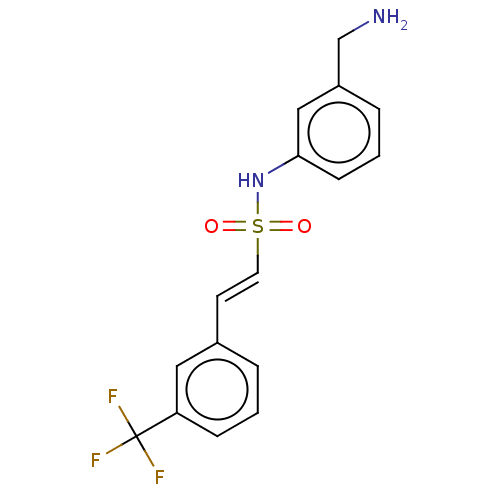
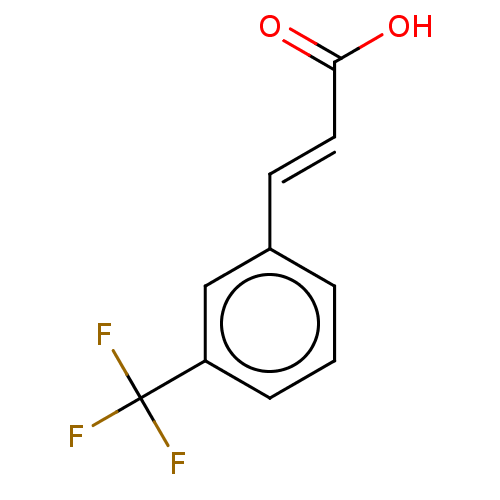
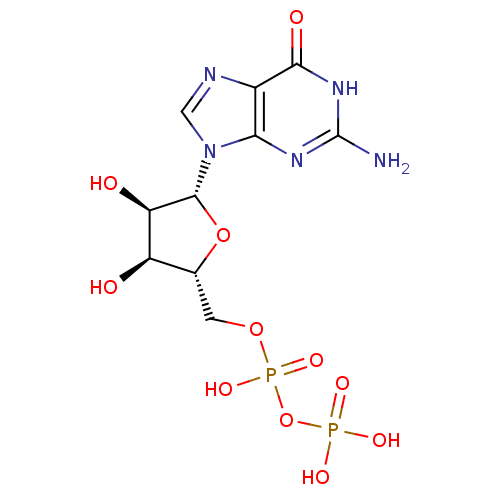
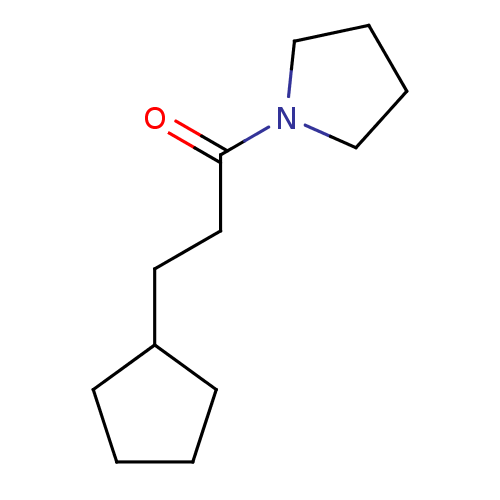
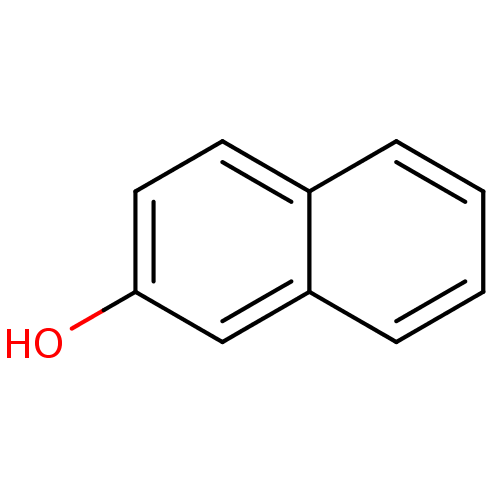
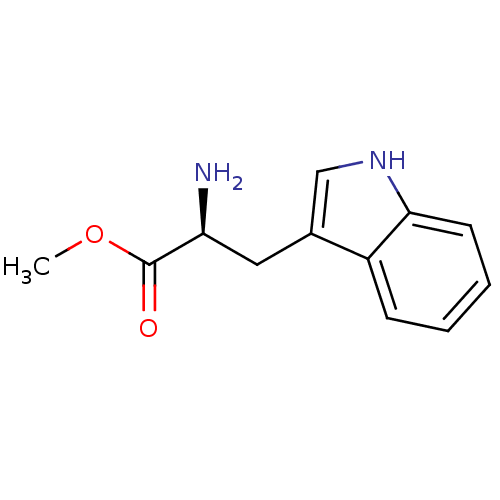
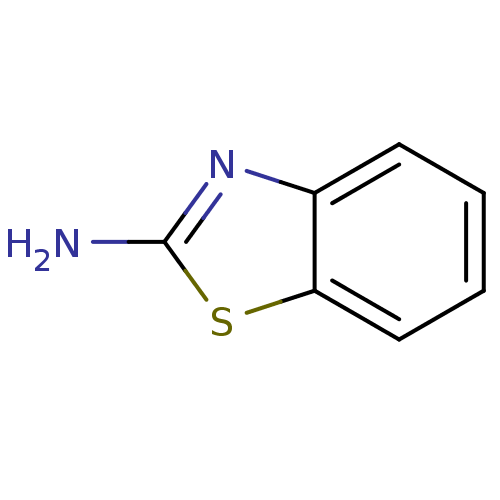
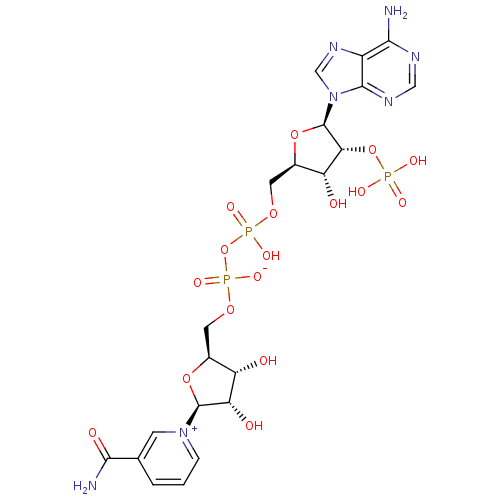
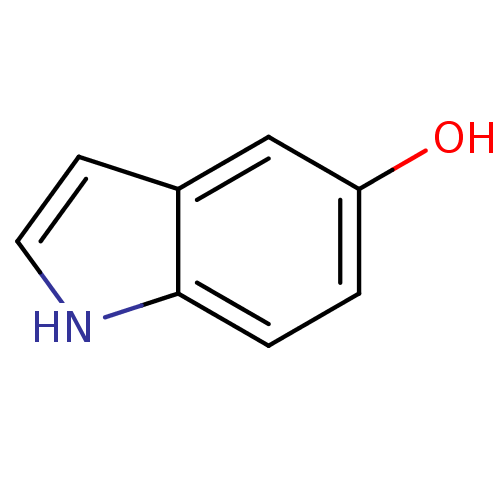
Target2-dehydropantoate 2-reductase(Escherichia coli (strain K12))
University Chemical Laboratory
Curated by ChEMBL
University Chemical Laboratory
Curated by ChEMBL
Affinity DataKi: 2.00E+5nMAssay Description:Inhibition of Escherichia coli KPRMore data for this Ligand-Target Pair
Target2-dehydropantoate 2-reductase(Escherichia coli (strain K12))
University Chemical Laboratory
Curated by ChEMBL
University Chemical Laboratory
Curated by ChEMBL
Affinity DataKi: 2.40E+5nMAssay Description:Inhibition of Escherichia coli KPRMore data for this Ligand-Target Pair
Target2-dehydropantoate 2-reductase(Escherichia coli (strain K12))
University Chemical Laboratory
Curated by ChEMBL
University Chemical Laboratory
Curated by ChEMBL
Affinity DataKi: 6.10E+5nMAssay Description:Inhibition of Escherichia coli KPRMore data for this Ligand-Target Pair
Target2-dehydropantoate 2-reductase(Escherichia coli (strain K12))
University Chemical Laboratory
Curated by ChEMBL
University Chemical Laboratory
Curated by ChEMBL
Affinity DataKi: 1.05E+6nMAssay Description:Inhibition of Escherichia coli KPRMore data for this Ligand-Target Pair
Target2-dehydropantoate 2-reductase(Escherichia coli (strain K12))
University Chemical Laboratory
Curated by ChEMBL
University Chemical Laboratory
Curated by ChEMBL
Affinity DataKi: 6.30E+6nMAssay Description:Inhibition of Escherichia coli KPRMore data for this Ligand-Target Pair
Target2-dehydropantoate 2-reductase(Escherichia coli (strain K12))
University Chemical Laboratory
Curated by ChEMBL
University Chemical Laboratory
Curated by ChEMBL
Affinity DataKi: 2.70E+7nMAssay Description:Inhibition of Escherichia coli KPRMore data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
University Of Cambridge
Curated by ChEMBL
University Of Cambridge
Curated by ChEMBL
Affinity DataIC50: 250nMAssay Description:Inhibition of hexahistidine-SUMO tagged Mycobacterium tuberculosis InhA expressed in Escherichia coli BL21(DE3) at preincubated for 10 mins with NADH...More data for this Ligand-Target Pair
TargetPantothenate synthetase(Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
Cambridge Crystallographic Data Centre
Curated by ChEMBL
Cambridge Crystallographic Data Centre
Curated by ChEMBL
Affinity DataIC50: 250nMAssay Description:Inhibition of Mycobacterium tuberculosis pantothenate synthetase expressed in Escherichia coliMore data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
University Of Cambridge
Curated by ChEMBL
University Of Cambridge
Curated by ChEMBL
Affinity DataIC50: 310nMAssay Description:Inhibition of hexahistidine-SUMO tagged Mycobacterium tuberculosis InhA expressed in Escherichia coli BL21(DE3) at preincubated for 10 mins with NADH...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
University Of Cambridge
Curated by ChEMBL
University Of Cambridge
Curated by ChEMBL
Affinity DataIC50: 4.00E+3nMAssay Description:Inhibition of hexahistidine-SUMO tagged Mycobacterium tuberculosis InhA expressed in Escherichia coli BL21(DE3) at preincubated for 10 mins with NADH...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
University Of Cambridge
Curated by ChEMBL
University Of Cambridge
Curated by ChEMBL
Affinity DataIC50: 6.00E+3nMAssay Description:Inhibition of hexahistidine-SUMO tagged Mycobacterium tuberculosis InhA expressed in Escherichia coli BL21(DE3) at preincubated for 10 mins with NADH...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
University Of Cambridge
Curated by ChEMBL
University Of Cambridge
Curated by ChEMBL
Affinity DataIC50: 9.00E+3nMAssay Description:Inhibition of hexahistidine-SUMO tagged Mycobacterium tuberculosis InhA expressed in Escherichia coli BL21(DE3) at preincubated for 10 mins with NADH...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
University Of Cambridge
Curated by ChEMBL
University Of Cambridge
Curated by ChEMBL
Affinity DataIC50: 2.20E+4nMAssay Description:Inhibition of hexahistidine-SUMO tagged Mycobacterium tuberculosis InhA expressed in Escherichia coli BL21(DE3) at preincubated for 10 mins with NADH...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
University Of Cambridge
Curated by ChEMBL
University Of Cambridge
Curated by ChEMBL
Affinity DataIC50: >1.00E+6nMAssay Description:Inhibition of hexahistidine-SUMO tagged Mycobacterium tuberculosis InhA expressed in Escherichia coli BL21(DE3) at preincubated for 10 mins with NADH...More data for this Ligand-Target Pair
Affinity DataKd: 2.00E+4nMAssay Description:An aqueous solution of the fragment to be tested (1 mM) was prepared containing NaCl (300 mM), Tris.HCl (20 mM, pH = 8.0), d6-DMSO (10% v/v) and glyc...More data for this Ligand-Target Pair
Affinity DataKd: 2.20E+4nMAssay Description:An aqueous solution of the fragment to be tested (1 mM) was prepared containing NaCl (300 mM), Tris.HCl (20 mM, pH = 8.0), d6-DMSO (10% v/v) and glyc...More data for this Ligand-Target Pair
Target2-dehydropantoate 2-reductase(Escherichia coli (strain K12))
University Chemical Laboratory
Curated by ChEMBL
University Chemical Laboratory
Curated by ChEMBL
Affinity DataKd: 1.53E+5nMAssay Description:Binding affinity to Escherichia coli KPRMore data for this Ligand-Target Pair
Target2-dehydropantoate 2-reductase(Escherichia coli (strain K12))
University Chemical Laboratory
Curated by ChEMBL
University Chemical Laboratory
Curated by ChEMBL
Affinity DataKd: 6.90E+6nMAssay Description:Binding affinity to Escherichia coli KPRMore data for this Ligand-Target Pair
Target2-dehydropantoate 2-reductase(Escherichia coli (strain K12))
University Chemical Laboratory
Curated by ChEMBL
University Chemical Laboratory
Curated by ChEMBL
Affinity DataKd: 6.30E+6nMAssay Description:Binding affinity to Escherichia coli KPRMore data for this Ligand-Target Pair
Target2-dehydropantoate 2-reductase(Escherichia coli (strain K12))
University Chemical Laboratory
Curated by ChEMBL
University Chemical Laboratory
Curated by ChEMBL
Affinity DataKd: 5.80E+3nMAssay Description:Binding affinity to Escherichia coli KPRMore data for this Ligand-Target Pair
Target2-dehydropantoate 2-reductase(Escherichia coli (strain K12))
University Chemical Laboratory
Curated by ChEMBL
University Chemical Laboratory
Curated by ChEMBL
Affinity DataKd: 6.10E+4nMAssay Description:Binding affinity to Escherichia coli KPRMore data for this Ligand-Target Pair
Target2-dehydropantoate 2-reductase(Escherichia coli (strain K12))
University Chemical Laboratory
Curated by ChEMBL
University Chemical Laboratory
Curated by ChEMBL
Affinity DataKd: >5.00E+6nMAssay Description:Binding affinity to Escherichia coli KPRMore data for this Ligand-Target Pair
Target2-dehydropantoate 2-reductase(Escherichia coli (strain K12))
University Chemical Laboratory
Curated by ChEMBL
University Chemical Laboratory
Curated by ChEMBL
Affinity DataKd: 6.60E+4nMAssay Description:Binding affinity to Escherichia coli KPRMore data for this Ligand-Target Pair
Target2-dehydropantoate 2-reductase(Escherichia coli (strain K12))
University Chemical Laboratory
Curated by ChEMBL
University Chemical Laboratory
Curated by ChEMBL
Affinity DataKd: 3.73E+5nMAssay Description:Binding affinity to Escherichia coli KPRMore data for this Ligand-Target Pair
Target2-dehydropantoate 2-reductase(Escherichia coli (strain K12))
University Chemical Laboratory
Curated by ChEMBL
University Chemical Laboratory
Curated by ChEMBL
Affinity DataKd: 1.00E+5nMAssay Description:Binding affinity to Escherichia coli KPRMore data for this Ligand-Target Pair
Target2-dehydropantoate 2-reductase(Escherichia coli (strain K12))
University Chemical Laboratory
Curated by ChEMBL
University Chemical Laboratory
Curated by ChEMBL
Affinity DataKd: 3.60E+5nMAssay Description:Binding affinity to Escherichia coli KPRMore data for this Ligand-Target Pair
Target2-dehydropantoate 2-reductase(Escherichia coli (strain K12))
University Chemical Laboratory
Curated by ChEMBL
University Chemical Laboratory
Curated by ChEMBL
Affinity DataKd: 1.30E+5nMAssay Description:Binding affinity to Escherichia coli KPRMore data for this Ligand-Target Pair
Affinity DataKd: 2.00E+4nMAssay Description:An aqueous solution of the fragment to be tested (1 mM) was prepared containing NaCl (300 mM), Tris.HCl (20 mM, pH = 8.0), d6-DMSO (10% v/v) and glyc...More data for this Ligand-Target Pair
Affinity DataKd: 1.60E+4nMAssay Description:An aqueous solution of the fragment to be tested (1 mM) was prepared containing NaCl (300 mM), Tris.HCl (20 mM, pH = 8.0), d6-DMSO (10% v/v) and glyc...More data for this Ligand-Target Pair
Affinity DataKd: 1.10E+4nMAssay Description:An aqueous solution of the fragment to be tested (1 mM) was prepared containing NaCl (300 mM), Tris.HCl (20 mM, pH = 8.0), d6-DMSO (10% v/v) and glyc...More data for this Ligand-Target Pair
Affinity DataKd: 6.00E+3nMAssay Description:An aqueous solution of the fragment to be tested (1 mM) was prepared containing NaCl (300 mM), Tris.HCl (20 mM, pH = 8.0), d6-DMSO (10% v/v) and glyc...More data for this Ligand-Target Pair
Affinity DataKd: 1.50E+4nMAssay Description:An aqueous solution of the fragment to be tested (1 mM) was prepared containing NaCl (300 mM), Tris.HCl (20 mM, pH = 8.0), d6-DMSO (10% v/v) and glyc...More data for this Ligand-Target Pair
Affinity DataKd: 1.20E+4nMAssay Description:An aqueous solution of the fragment to be tested (1 mM) was prepared containing NaCl (300 mM), Tris.HCl (20 mM, pH = 8.0), d6-DMSO (10% v/v) and glyc...More data for this Ligand-Target Pair
Affinity DataKd: 4.60E+5nMAssay Description:The thermal-shift denaturation assay was performed on an iCycler iQ Real Time Detection System (BioRad) in 96-well iCycler iQ PCR plates sealed with ...More data for this Ligand-Target Pair
Affinity DataKd: 4.30E+5nMAssay Description:The thermal-shift denaturation assay was performed on an iCycler iQ Real Time Detection System (BioRad) in 96-well iCycler iQ PCR plates sealed with ...More data for this Ligand-Target Pair
Affinity DataKd: 5.70E+5nMAssay Description:The thermal-shift denaturation assay was performed on an iCycler iQ Real Time Detection System (BioRad) in 96-well iCycler iQ PCR plates sealed with ...More data for this Ligand-Target Pair
Affinity DataKd: 7.30E+5nMAssay Description:The thermal-shift denaturation assay was performed on an iCycler iQ Real Time Detection System (BioRad) in 96-well iCycler iQ PCR plates sealed with ...More data for this Ligand-Target Pair
Affinity DataKd: 1.00E+6nMAssay Description:The thermal-shift denaturation assay was performed on an iCycler iQ Real Time Detection System (BioRad) in 96-well iCycler iQ PCR plates sealed with ...More data for this Ligand-Target Pair
Affinity DataKd: 1.30E+6nMAssay Description:The thermal-shift denaturation assay was performed on an iCycler iQ Real Time Detection System (BioRad) in 96-well iCycler iQ PCR plates sealed with ...More data for this Ligand-Target Pair
Affinity DataKd: 2.10E+6nMAssay Description:The thermal-shift denaturation assay was performed on an iCycler iQ Real Time Detection System (BioRad) in 96-well iCycler iQ PCR plates sealed with ...More data for this Ligand-Target Pair
Target2-dehydropantoate 2-reductase(Escherichia coli (strain K12))
University Chemical Laboratory
Curated by ChEMBL
University Chemical Laboratory
Curated by ChEMBL
Affinity DataKd: 5.80E+3nMAssay Description:Binding affinity to Escherichia coli KPRMore data for this Ligand-Target Pair
Affinity DataKd: 2.10E+6nMAssay Description:The thermal-shift denaturation assay was performed on an iCycler iQ Real Time Detection System (BioRad) in 96-well iCycler iQ PCR plates sealed with ...More data for this Ligand-Target Pair

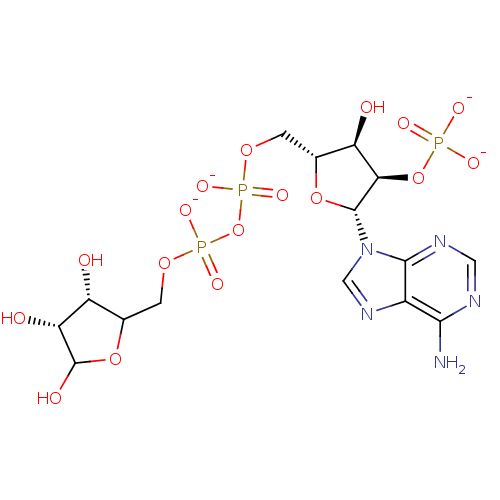


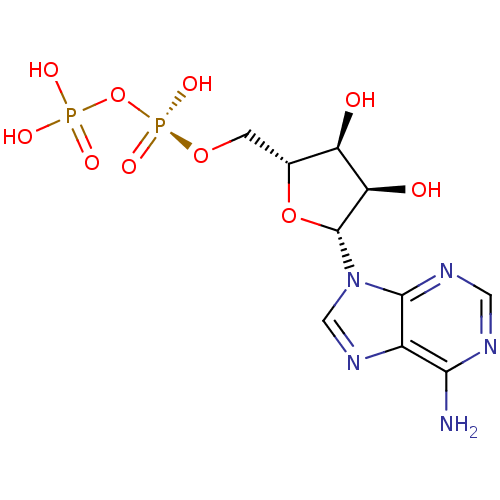
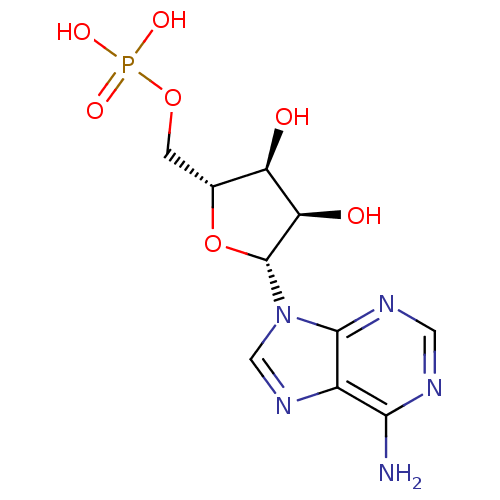

 3D Structure (crystal)
3D Structure (crystal)