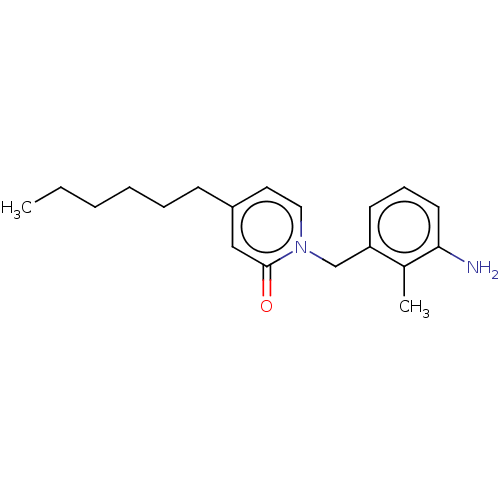
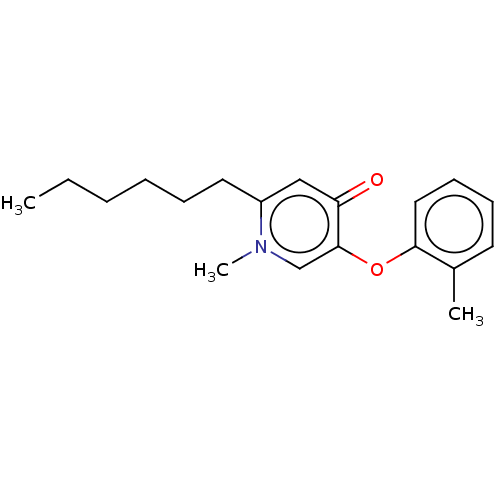
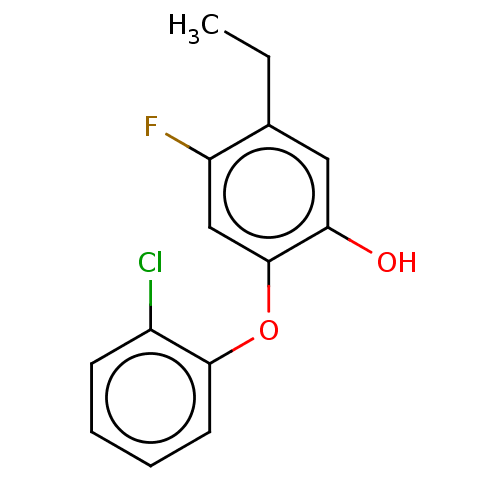
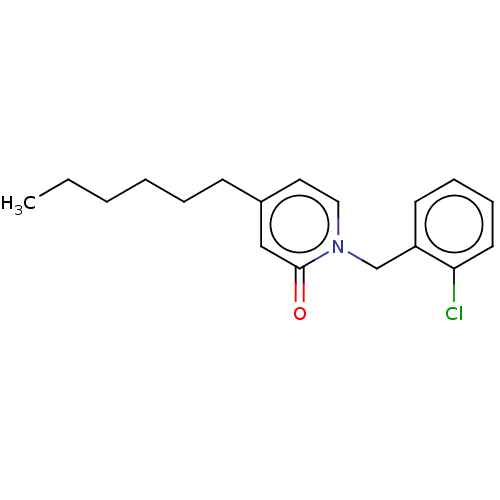
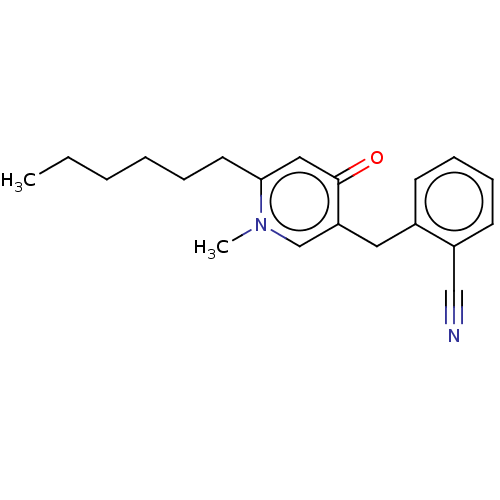
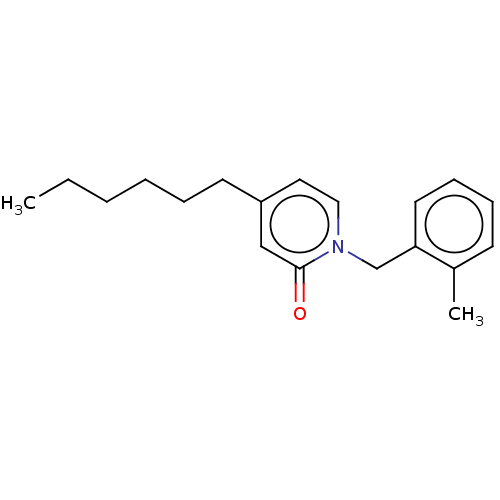
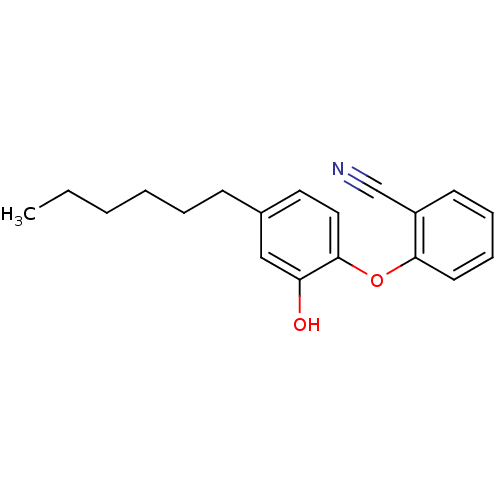
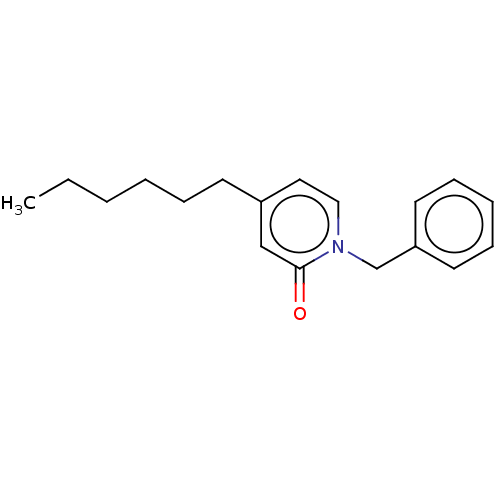
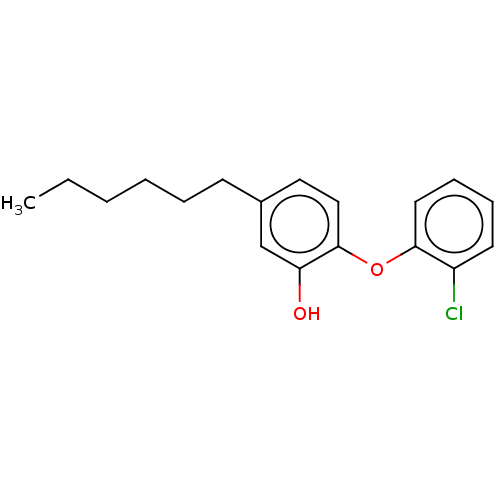
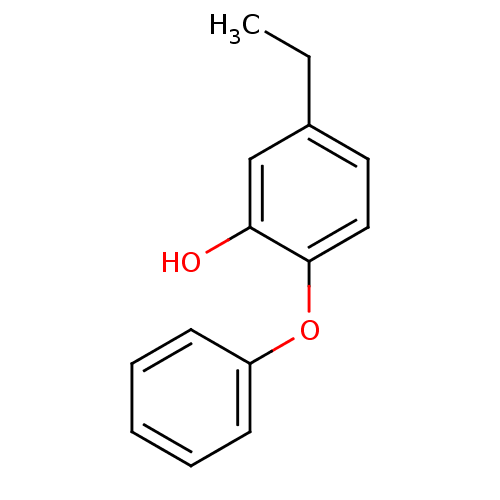
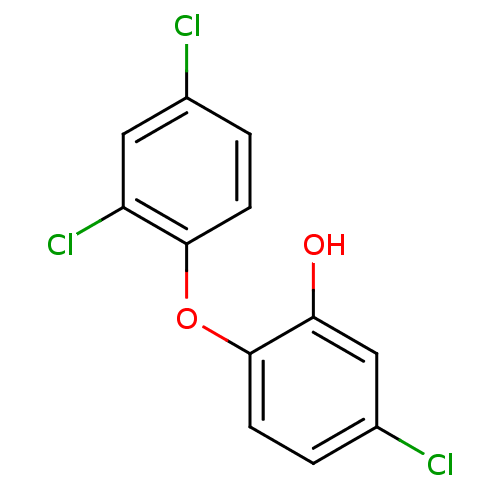
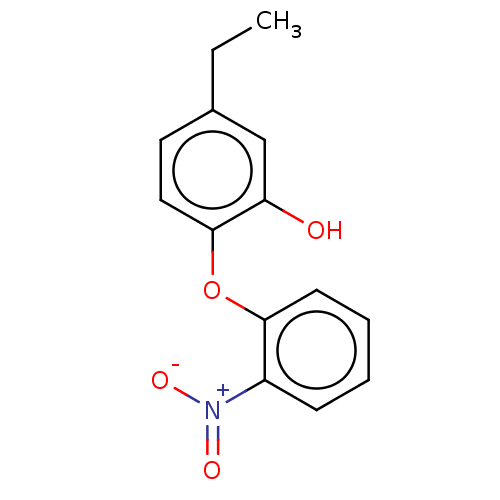
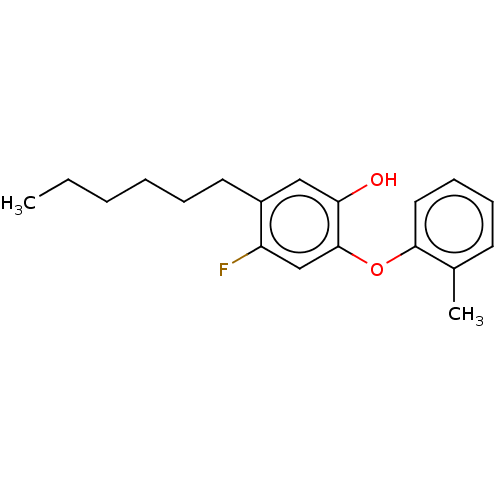
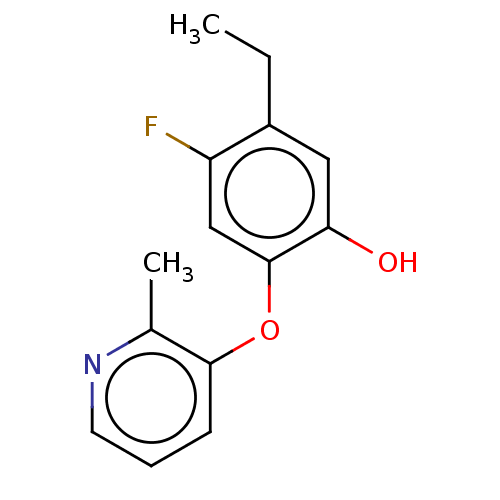
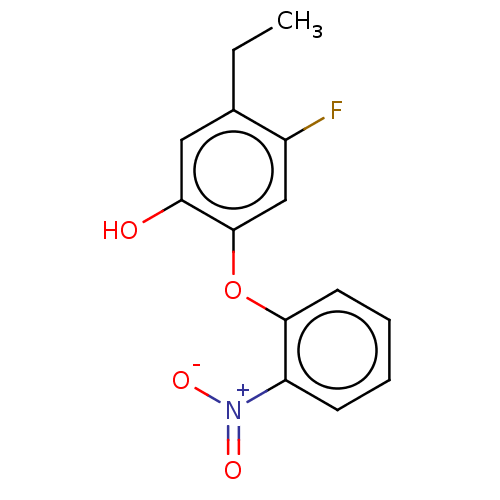
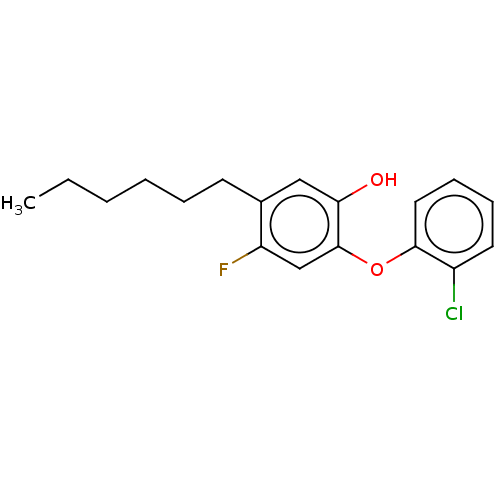
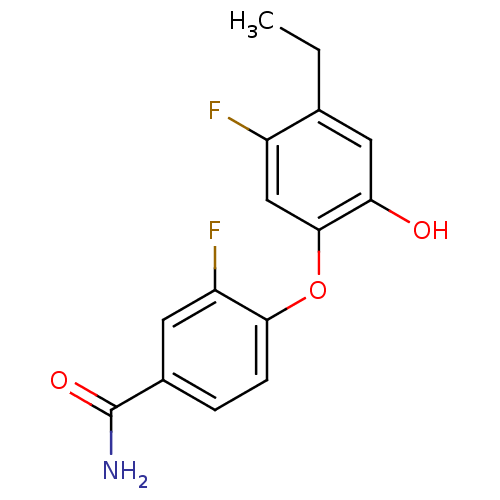
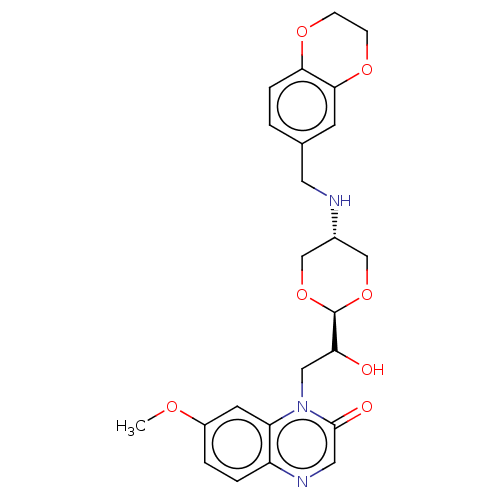
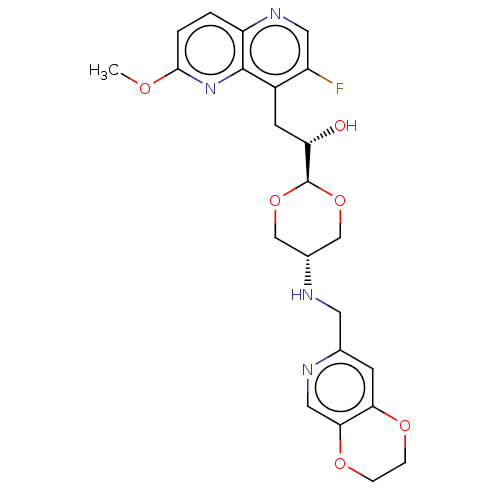
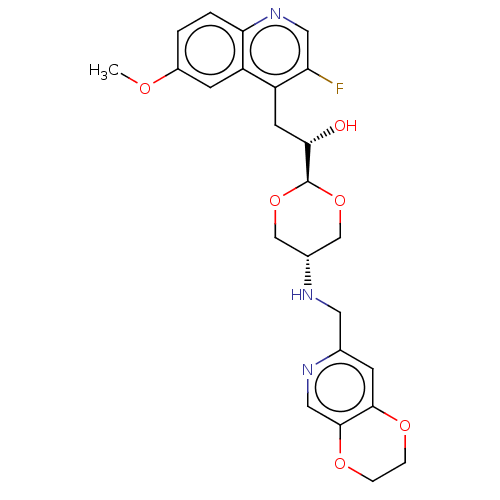
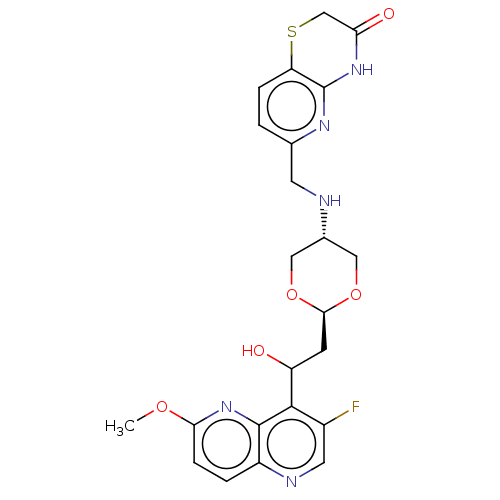
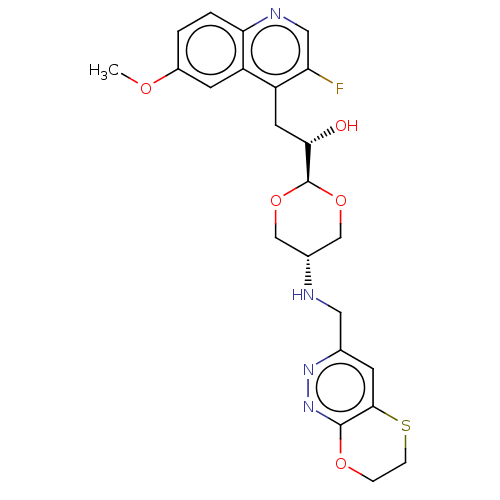
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Burkholderia pseudomallei)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.510nM ΔG°: -53.0kJ/molepH: 8.0 T: 2°CAssay Description:Slow-onset inhibition kinetics were monitored at 340 nm on a Cary 100 spectrophotometer (Varian) at 25 °C in 30 mM PIPES buffer (pH 8.0) containing 1...More data for this Ligand-Target Pair
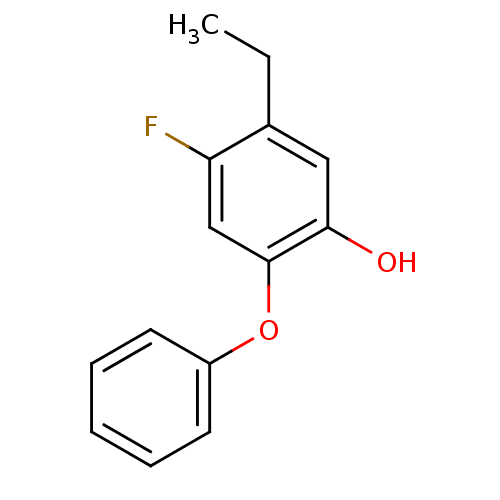
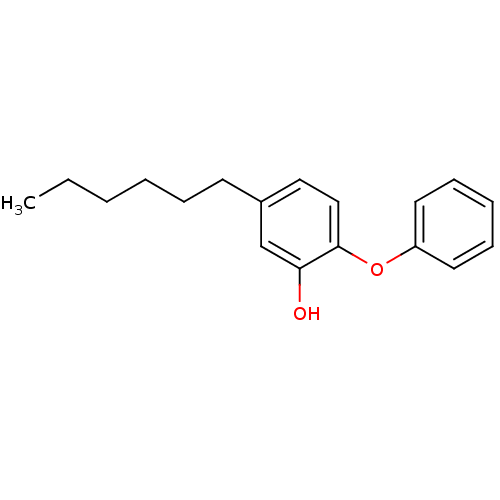
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Burkholderia pseudomallei)
Stony Brook University
Stony Brook University
Affinity DataKi: 1nM ΔG°: -51.4kJ/molepH: 8.0 T: 2°CAssay Description:Slow-onset inhibition kinetics were monitored at 340 nm on a Cary 100 spectrophotometer (Varian) at 25 °C in 30 mM PIPES buffer (pH 8.0) containing 1...More data for this Ligand-Target Pair
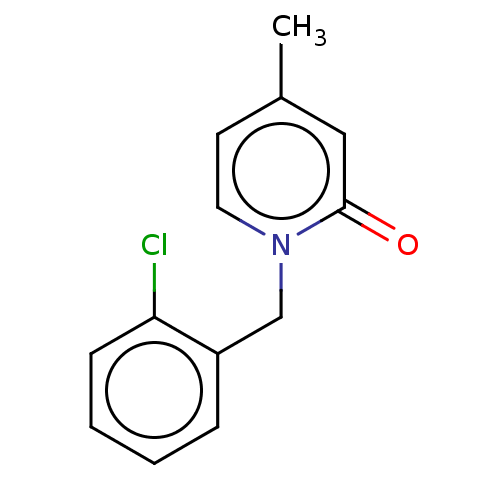
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
University Of Wuerzburg
University Of Wuerzburg
Affinity DataKi: 1.30nMpH: 7.5Assay Description:Kinetics were performed on a Cary 100 spectrophotometer (Varian) at 20 °C. Reaction velocities were measured by monitoring the oxidation of NAD(P)H t...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
University Of Wuerzburg
University Of Wuerzburg
Affinity DataKi: 2nMpH: 7.5Assay Description:Kinetics were performed on a Cary 100 spectrophotometer (Varian) at 20 °C. Reaction velocities were measured by monitoring the oxidation of NAD(P)H t...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
University Of Wuerzburg
University Of Wuerzburg
Affinity DataKi: 2.70nMpH: 7.5Assay Description:Kinetics were performed on a Cary 100 spectrophotometer (Varian) at 20 °C. Reaction velocities were measured by monitoring the oxidation of NAD(P)H t...More data for this Ligand-Target Pair
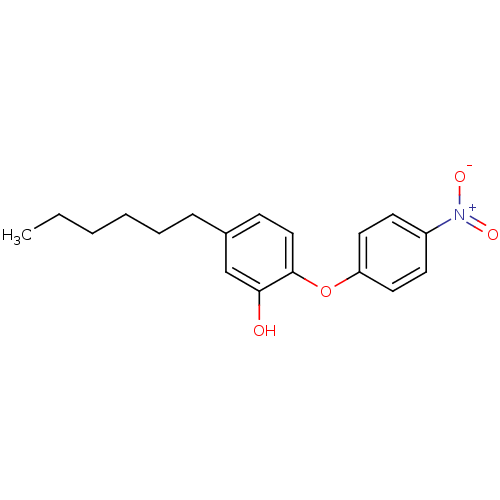
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Burkholderia pseudomallei)
Stony Brook University
Stony Brook University
Affinity DataKi: 3nM ΔG°: -48.6kJ/molepH: 8.0 T: 2°CAssay Description:Slow-onset inhibition kinetics were monitored at 340 nm on a Cary 100 spectrophotometer (Varian) at 25 °C in 30 mM PIPES buffer (pH 8.0) containing 1...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Burkholderia pseudomallei)
Stony Brook University
Stony Brook University
Affinity DataKi: 4nM ΔG°: -47.9kJ/molepH: 8.0 T: 2°CAssay Description:Slow-onset inhibition kinetics were monitored at 340 nm on a Cary 100 spectrophotometer (Varian) at 25 °C in 30 mM PIPES buffer (pH 8.0) containing 1...More data for this Ligand-Target Pair
Affinity DataKi: 7nMpH: 7.5Assay Description:Kinetics were performed on a Cary 100 spectrophotometer (Varian) at 20 °C. Reaction velocities were measured by monitoring the oxidation of NAD(P)H t...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
University Of Wuerzburg
University Of Wuerzburg
Affinity DataKi: 11nMpH: 7.5Assay Description:Kinetics were performed on a Cary 100 spectrophotometer (Varian) at 20 °C. Reaction velocities were measured by monitoring the oxidation of NAD(P)H t...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
University Of Wuerzburg
University Of Wuerzburg
Affinity DataKi: 11nMpH: 7.5Assay Description:Kinetics were performed on a Cary 100 spectrophotometer (Varian) at 20 °C. Reaction velocities were measured by monitoring the oxidation of NAD(P)H t...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
University Of Wuerzburg
University Of Wuerzburg
Affinity DataKi: 11.9nMpH: 7.5Assay Description:Kinetics were performed on a Cary 100 spectrophotometer (Varian) at 20 °C. Reaction velocities were measured by monitoring the oxidation of NAD(P)H t...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Burkholderia pseudomallei)
Stony Brook University
Stony Brook University
Affinity DataKi: 15nM ΔG°: -44.7kJ/molepH: 8.0 T: 2°CAssay Description:Slow-onset inhibition kinetics were monitored at 340 nm on a Cary 100 spectrophotometer (Varian) at 25 °C in 30 mM PIPES buffer (pH 8.0) containing 1...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
University Of Wuerzburg
University Of Wuerzburg
Affinity DataKi: 19nMpH: 7.5Assay Description:Kinetics were performed on a Cary 100 spectrophotometer (Varian) at 20 °C. Reaction velocities were measured by monitoring the oxidation of NAD(P)H t...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Burkholderia pseudomallei)
Stony Brook University
Stony Brook University
Affinity DataKi: 25nM ΔG°: -43.4kJ/molepH: 8.0 T: 2°CAssay Description:Slow-onset inhibition kinetics were monitored at 340 nm on a Cary 100 spectrophotometer (Varian) at 25 °C in 30 mM PIPES buffer (pH 8.0) containing 1...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
University Of Wuerzburg
University Of Wuerzburg
Affinity DataKi: 29.6nMpH: 7.5Assay Description:Kinetics were performed on a Cary 100 spectrophotometer (Varian) at 20 °C. Reaction velocities were measured by monitoring the oxidation of NAD(P)H t...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Burkholderia pseudomallei)
Stony Brook University
Stony Brook University
Affinity DataKi: 31nM ΔG°: -42.9kJ/molepH: 8.0 T: 2°CAssay Description:Slow-onset inhibition kinetics were monitored at 340 nm on a Cary 100 spectrophotometer (Varian) at 25 °C in 30 mM PIPES buffer (pH 8.0) containing 1...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Burkholderia pseudomallei)
Stony Brook University
Stony Brook University
Affinity DataKi: 32nM ΔG°: -42.8kJ/molepH: 8.0 T: 2°CAssay Description:Slow-onset inhibition kinetics were monitored at 340 nm on a Cary 100 spectrophotometer (Varian) at 25 °C in 30 mM PIPES buffer (pH 8.0) containing 1...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Burkholderia pseudomallei)
Stony Brook University
Stony Brook University
Affinity DataKi: 32nM ΔG°: -42.8kJ/molepH: 8.0 T: 2°CAssay Description:Slow-onset inhibition kinetics were monitored at 340 nm on a Cary 100 spectrophotometer (Varian) at 25 °C in 30 mM PIPES buffer (pH 8.0) containing 1...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Burkholderia pseudomallei)
Stony Brook University
Stony Brook University
Affinity DataKi: 32nM ΔG°: -42.8kJ/molepH: 8.0 T: 2°CAssay Description:Slow-onset inhibition kinetics were monitored at 340 nm on a Cary 100 spectrophotometer (Varian) at 25 °C in 30 mM PIPES buffer (pH 8.0) containing 1...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Burkholderia pseudomallei)
Stony Brook University
Stony Brook University
Affinity DataKi: 44nM ΔG°: -42.0kJ/molepH: 8.0 T: 2°CAssay Description:Slow-onset inhibition kinetics were monitored at 340 nm on a Cary 100 spectrophotometer (Varian) at 25 °C in 30 mM PIPES buffer (pH 8.0) containing 1...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Burkholderia pseudomallei)
Stony Brook University
Stony Brook University
Affinity DataKi: 48nM ΔG°: -41.8kJ/molepH: 8.0 T: 2°CAssay Description:Slow-onset inhibition kinetics were monitored at 340 nm on a Cary 100 spectrophotometer (Varian) at 25 °C in 30 mM PIPES buffer (pH 8.0) containing 1...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Burkholderia pseudomallei)
Stony Brook University
Stony Brook University
Affinity DataKi: 55nM ΔG°: -41.4kJ/molepH: 8.0 T: 2°CAssay Description:Slow-onset inhibition kinetics were monitored at 340 nm on a Cary 100 spectrophotometer (Varian) at 25 °C in 30 mM PIPES buffer (pH 8.0) containing 1...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Burkholderia pseudomallei)
Stony Brook University
Stony Brook University
Affinity DataKi: 55nM ΔG°: -41.4kJ/molepH: 8.0 T: 2°CAssay Description:Slow-onset inhibition kinetics were monitored at 340 nm on a Cary 100 spectrophotometer (Varian) at 25 °C in 30 mM PIPES buffer (pH 8.0) containing 1...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Burkholderia pseudomallei)
Stony Brook University
Stony Brook University
Affinity DataKi: 58nM ΔG°: -41.3kJ/molepH: 8.0 T: 2°CAssay Description:Slow-onset inhibition kinetics were monitored at 340 nm on a Cary 100 spectrophotometer (Varian) at 25 °C in 30 mM PIPES buffer (pH 8.0) containing 1...More data for this Ligand-Target Pair
Affinity DataKi: 81.9nMpH: 7.5Assay Description:Kinetics were performed on a Cary 100 spectrophotometer (Varian) at 20 °C. Reaction velocities were measured by monitoring the oxidation of NAD(P)H t...More data for this Ligand-Target Pair
Affinity DataKi: 95.7nMpH: 7.5Assay Description:Kinetics were performed on a Cary 100 spectrophotometer (Varian) at 20 °C. Reaction velocities were measured by monitoring the oxidation of NAD(P)H t...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Burkholderia pseudomallei)
Stony Brook University
Stony Brook University
Affinity DataKi: 105nM ΔG°: -39.8kJ/molepH: 8.0 T: 2°CAssay Description:Slow-onset inhibition kinetics were monitored at 340 nm on a Cary 100 spectrophotometer (Varian) at 25 °C in 30 mM PIPES buffer (pH 8.0) containing 1...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Burkholderia pseudomallei)
Stony Brook University
Stony Brook University
Affinity DataKi: 130nM ΔG°: -39.3kJ/molepH: 8.0 T: 2°CAssay Description:Slow-onset inhibition kinetics were monitored at 340 nm on a Cary 100 spectrophotometer (Varian) at 25 °C in 30 mM PIPES buffer (pH 8.0) containing 1...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Burkholderia pseudomallei)
Stony Brook University
Stony Brook University
Affinity DataKi: 138nM ΔG°: -39.2kJ/molepH: 8.0 T: 2°CAssay Description:Slow-onset inhibition kinetics were monitored at 340 nm on a Cary 100 spectrophotometer (Varian) at 25 °C in 30 mM PIPES buffer (pH 8.0) containing 1...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Burkholderia pseudomallei)
Stony Brook University
Stony Brook University
Affinity DataKi: 153nM ΔG°: -38.9kJ/molepH: 8.0 T: 2°CAssay Description:Slow-onset inhibition kinetics were monitored at 340 nm on a Cary 100 spectrophotometer (Varian) at 25 °C in 30 mM PIPES buffer (pH 8.0) containing 1...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Burkholderia pseudomallei)
Stony Brook University
Stony Brook University
Affinity DataKi: 156nM ΔG°: -38.9kJ/molepH: 8.0 T: 2°CAssay Description:Slow-onset inhibition kinetics were monitored at 340 nm on a Cary 100 spectrophotometer (Varian) at 25 °C in 30 mM PIPES buffer (pH 8.0) containing 1...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Burkholderia pseudomallei)
Stony Brook University
Stony Brook University
Affinity DataKi: 158nM ΔG°: -38.8kJ/molepH: 8.0 T: 2°CAssay Description:Slow-onset inhibition kinetics were monitored at 340 nm on a Cary 100 spectrophotometer (Varian) at 25 °C in 30 mM PIPES buffer (pH 8.0) containing 1...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
University Of Wuerzburg
University Of Wuerzburg
Affinity DataKi: 197nMpH: 7.5Assay Description:Kinetics were performed on a Cary 100 spectrophotometer (Varian) at 20 °C. Reaction velocities were measured by monitoring the oxidation of NAD(P)H t...More data for this Ligand-Target Pair
Affinity DataKi: 203nMpH: 7.5Assay Description:Kinetics were performed on a Cary 100 spectrophotometer (Varian) at 20 °C. Reaction velocities were measured by monitoring the oxidation of NAD(P)H t...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
University Of Wuerzburg
University Of Wuerzburg
Affinity DataKi: 229nMpH: 7.5Assay Description:Kinetics were performed on a Cary 100 spectrophotometer (Varian) at 20 °C. Reaction velocities were measured by monitoring the oxidation of NAD(P)H t...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Burkholderia pseudomallei)
Stony Brook University
Stony Brook University
Affinity DataKi: 405nM ΔG°: -36.5kJ/molepH: 8.0 T: 2°CAssay Description:Slow-onset inhibition kinetics were monitored at 340 nm on a Cary 100 spectrophotometer (Varian) at 25 °C in 30 mM PIPES buffer (pH 8.0) containing 1...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Burkholderia pseudomallei)
Stony Brook University
Stony Brook University
Affinity DataKi: 1.13E+3nM ΔG°: -33.9kJ/molepH: 8.0 T: 2°CAssay Description:Slow-onset inhibition kinetics were monitored at 340 nm on a Cary 100 spectrophotometer (Varian) at 25 °C in 30 mM PIPES buffer (pH 8.0) containing 1...More data for this Ligand-Target Pair
Affinity DataIC50: 13nMAssay Description:Inhibition of Staphylococcus aureus Gyrase supercoiling activity assessed as reduction in supercoiling pBR322 DNA by measuring relaxation by bromophe...More data for this Ligand-Target Pair
Affinity DataIC50: 14nMAssay Description:Inhibition of Staphylococcus aureus Gyrase supercoiling activity assessed as reduction in supercoiling pBR322 DNA by measuring relaxation by bromophe...More data for this Ligand-Target Pair
Affinity DataIC50: 19nMAssay Description:Inhibition of Staphylococcus aureus Gyrase supercoiling activity assessed as reduction in supercoiling pBR322 DNA by measuring relaxation by bromophe...More data for this Ligand-Target Pair
Affinity DataIC50: 22nMAssay Description:Inhibition of Staphylococcus aureus Gyrase supercoiling activity assessed as reduction in supercoiling pBR322 DNA by measuring relaxation by bromophe...More data for this Ligand-Target Pair
Affinity DataIC50: 31nMAssay Description:Inhibition of Staphylococcus aureus Gyrase supercoiling activity assessed as reduction in supercoiling pBR322 DNA by measuring relaxation by bromophe...More data for this Ligand-Target Pair
Affinity DataIC50: 39nMAssay Description:Inhibition of Staphylococcus aureus Gyrase supercoiling activity assessed as reduction in supercoiling pBR322 DNA by measuring relaxation by bromophe...More data for this Ligand-Target Pair
Affinity DataIC50: 42nMAssay Description:Inhibition of Staphylococcus aureus Gyrase supercoiling activity assessed as reduction in supercoiling pBR322 DNA by measuring relaxation by bromophe...More data for this Ligand-Target Pair
Affinity DataIC50: 43nMAssay Description:Inhibition of Staphylococcus aureus Gyrase supercoiling activity assessed as reduction in supercoiling pBR322 DNA by measuring relaxation by bromophe...More data for this Ligand-Target Pair
Affinity DataIC50: 54nMAssay Description:Inhibition of Staphylococcus aureus Gyrase supercoiling activity assessed as reduction in supercoiling pBR322 DNA by measuring relaxation by bromophe...More data for this Ligand-Target Pair
Affinity DataIC50: 56nMAssay Description:Inhibition of Staphylococcus aureus Gyrase supercoiling activity assessed as reduction in supercoiling pBR322 DNA by measuring relaxation by bromophe...More data for this Ligand-Target Pair
Affinity DataIC50: 56nMAssay Description:Inhibition of Staphylococcus aureus Gyrase supercoiling activity assessed as reduction in supercoiling pBR322 DNA by measuring relaxation by bromophe...More data for this Ligand-Target Pair
Affinity DataIC50: 60nMAssay Description:Inhibition of Staphylococcus aureus Gyrase supercoiling activity assessed as reduction in supercoiling pBR322 DNA by measuring relaxation by bromophe...More data for this Ligand-Target Pair
Affinity DataIC50: 61nMAssay Description:Inhibition of Staphylococcus aureus Gyrase supercoiling activity assessed as reduction in supercoiling pBR322 DNA by measuring relaxation by bromophe...More data for this Ligand-Target Pair


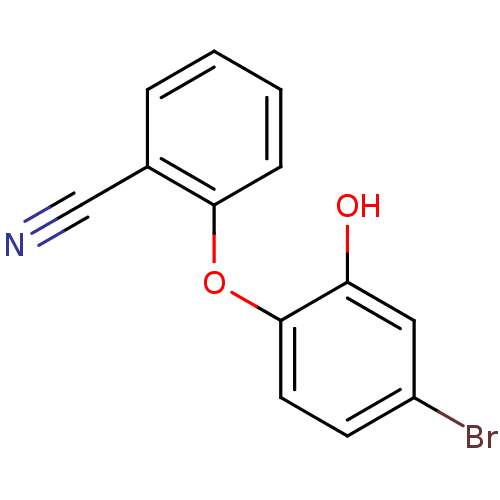
 3D Structure (crystal)
3D Structure (crystal)