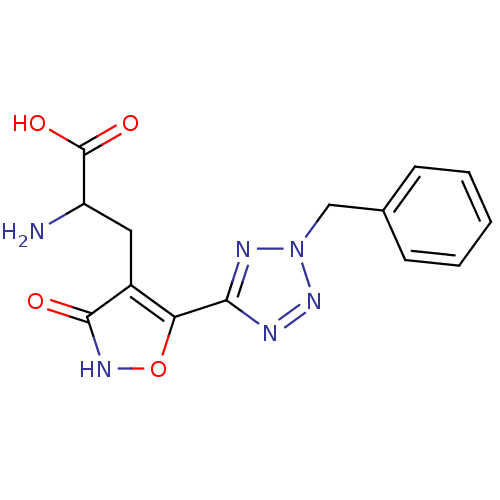
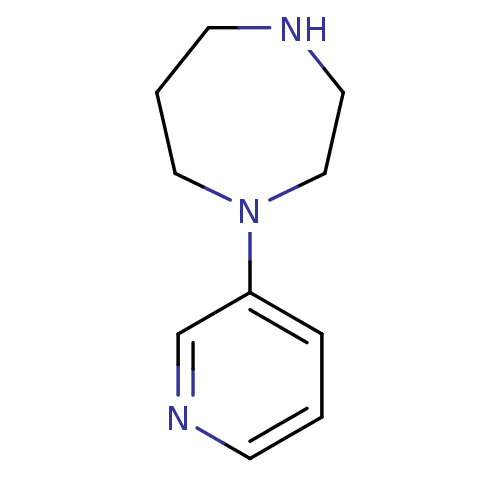
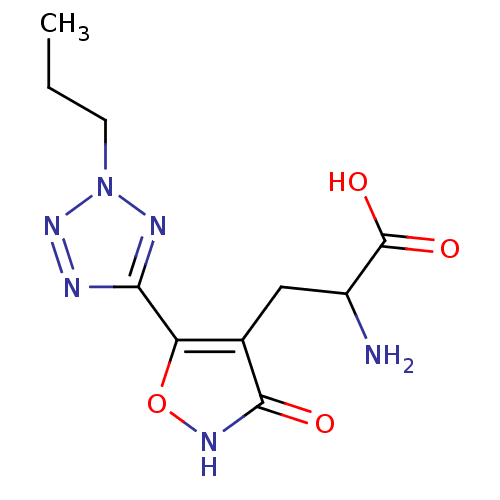
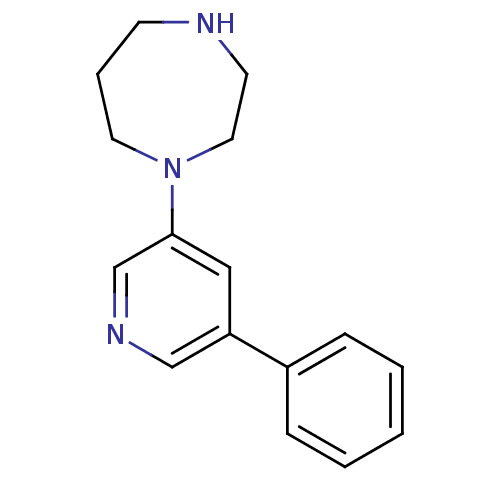
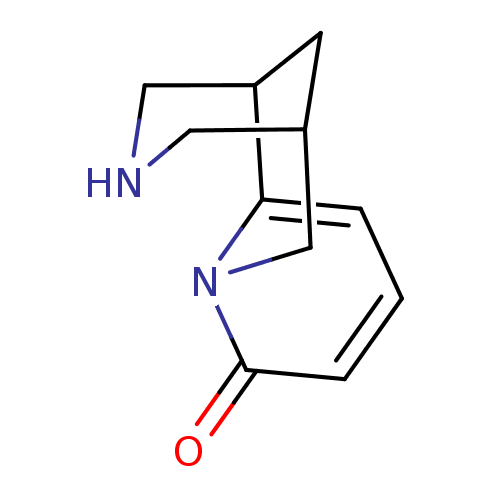
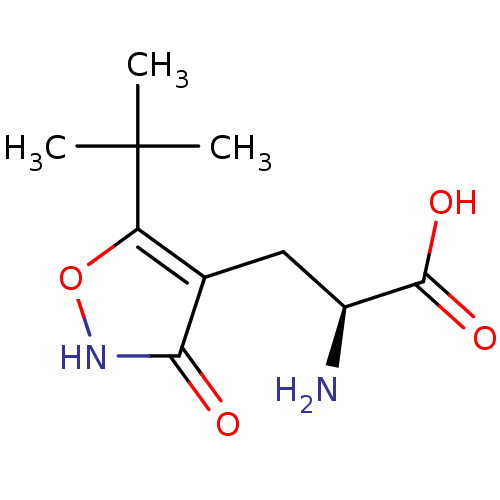
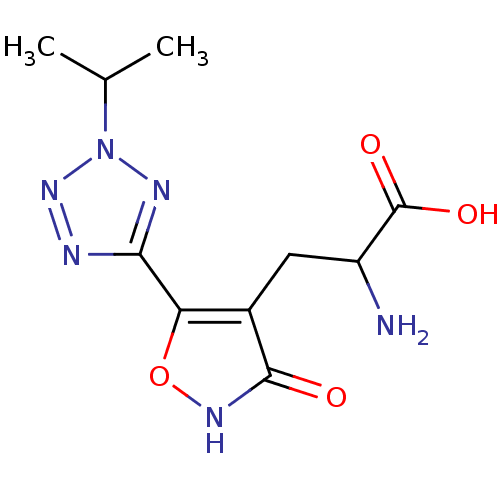
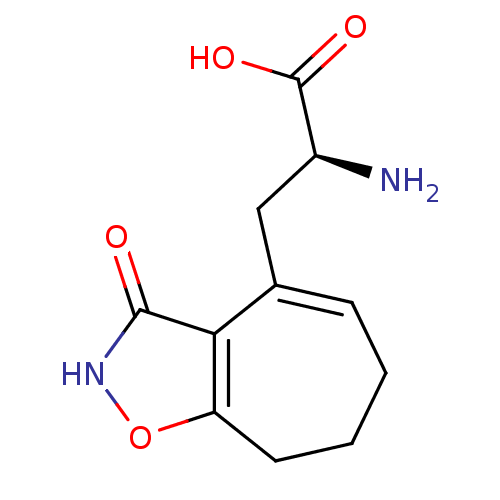
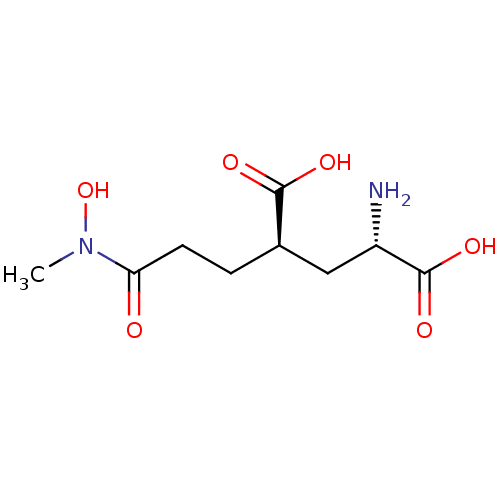
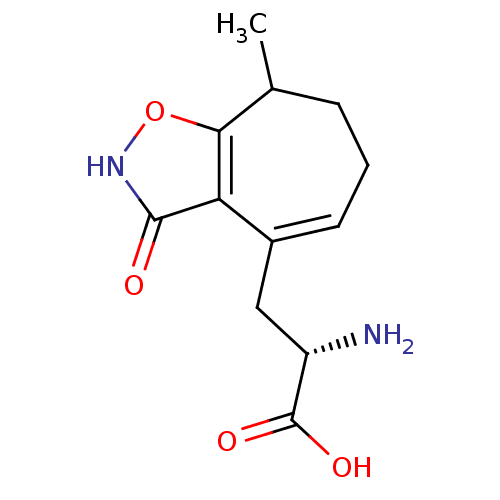
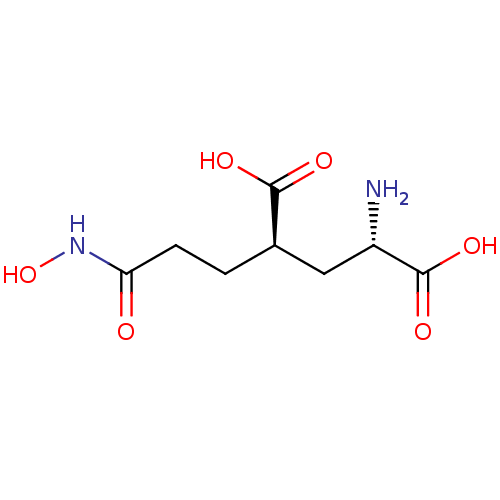
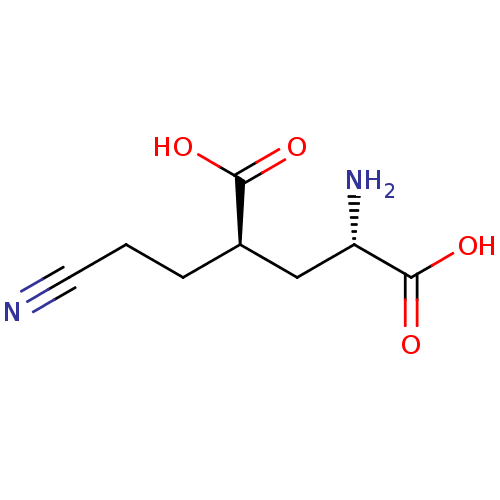
Affinity DataKi: 0.00220nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR3 expressed in Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 0.00270nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR4 expressed in Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 0.00390nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR2 expressed in Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 0.00520nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR1 expressed in Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 0.0170nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR2 expressed in Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 0.0210nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR3 expressed in Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 0.0220nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR1 expressed in Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 0.0400nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR4 expressed in Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 0.0440nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR4 expressed in Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 0.0450nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR3 expressed in Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 0.0670nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
Affinity DataKi: 0.0800nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR2 expressed in Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 0.0810nMAssay Description:Displacement of [3H]SYM 2081 from rat recombinant GluR5 expressed in Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 0.0970nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
Affinity DataKi: 0.150nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR1 expressed in Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 0.200nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR4 expressed in Sf9 cellsMore data for this Ligand-Target Pair
TargetNeuronal acetylcholine receptor subunit alpha-4(Rattus norvegicus (Rat))
University Of Copenhagen
University Of Copenhagen
Affinity DataKi: 0.25nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
TargetNeuronal acetylcholine receptor subunit alpha-4(Rattus norvegicus (Rat))
University Of Copenhagen
University Of Copenhagen
Affinity DataKi: 0.320nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
TargetNeuronal acetylcholine receptor subunit alpha-4(Rattus norvegicus (Rat))
University Of Copenhagen
University Of Copenhagen
Affinity DataKi: 0.330nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
Affinity DataKi: 0.390nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR3 expressed in Sf9 cellsMore data for this Ligand-Target Pair
TargetNeuronal acetylcholine receptor subunit alpha-4(Rattus norvegicus (Rat))
University Of Copenhagen
University Of Copenhagen
Affinity DataKi: 0.620nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
TargetNeuronal acetylcholine receptor subunit alpha-4(Rattus norvegicus (Rat))
University Of Copenhagen
University Of Copenhagen
Affinity DataKi: 0.720nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
Affinity DataKi: 0.730nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR4 expressed in Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 0.75nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR2 expressed in Sf9 cellsMore data for this Ligand-Target Pair
TargetNeuronal acetylcholine receptor subunit alpha-4(Rattus norvegicus (Rat))
University Of Copenhagen
University Of Copenhagen
Affinity DataKi: 0.800nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
TargetNeuronal acetylcholine receptor subunit alpha-4(Rattus norvegicus (Rat))
University Of Copenhagen
University Of Copenhagen
Affinity DataKi: 0.950nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
Affinity DataKi: 1.20nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR3 expressed in Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 1.20nMAssay Description:Displacement of [3H]SYM 2081 from rat recombinant GluR5 expressed in Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 1.30nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
Affinity DataKi: 1.60nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR2 expressed in Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 2.13nMAssay Description:Displacement of [3H]SYM from rat recombinant GluR5(Q) RNA-edited isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 2.20nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
Affinity DataKi: 2.40nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR4 expressed in Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 2.57nMAssay Description:Displacement of [3H]SYM from rat recombinant GluR5(Q) RNA-edited isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 2.90nMAssay Description:Displacement of [3H]SYM 2081 from rat recombinant GluR5 expressed in Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 2.90nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR2 expressed in Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 3.10nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
Affinity DataKi: 3.40nMAssay Description:Displacement of [3H]-(2S,4R)-4-methylglutamic acid from full length recombinant rat GluKK1(Q)1b receptor expressed in sf9 cells by liquid scintillati...More data for this Ligand-Target Pair
Affinity DataKi: 3.70nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR3 expressed in Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 5.08nMAssay Description:Displacement of [3H]SYM from rat recombinant GluR5(Q) RNA-edited isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 5.20nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR1 expressed in Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 5.5nMAssay Description:Displacement of [3H]SYM 2081 from rat recombinant GluR5 expressed in Sf9 cellsMore data for this Ligand-Target Pair
TargetNeuronal acetylcholine receptor subunit alpha-4(Rattus norvegicus (Rat))
University Of Copenhagen
University Of Copenhagen
Affinity DataKi: 6nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
Affinity DataKi: 7.70nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR1 expressed in Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 8.20nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
Affinity DataKi: 8.90nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
Affinity DataKi: 10nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR1 expressed in Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 13nMAssay Description:Displacement of [3H]-(2S,4R)-4-methylglutamic acid from full length recombinant rat GluKK1(Q)1b receptor expressed in sf9 cells by liquid scintillati...More data for this Ligand-Target Pair
Affinity DataKi: 17nMAssay Description:Displacement of [3H]-(2S,4R)-4-methylglutamic acid from full length recombinant rat GluKK1(Q)1b receptor expressed in sf9 cells by liquid scintillati...More data for this Ligand-Target Pair
TargetNeuronal acetylcholine receptor subunit alpha-7(Rattus norvegicus (Rat))
University Of Copenhagen
University Of Copenhagen
Affinity DataKi: 18nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair


 3D Structure (crystal)
3D Structure (crystal)