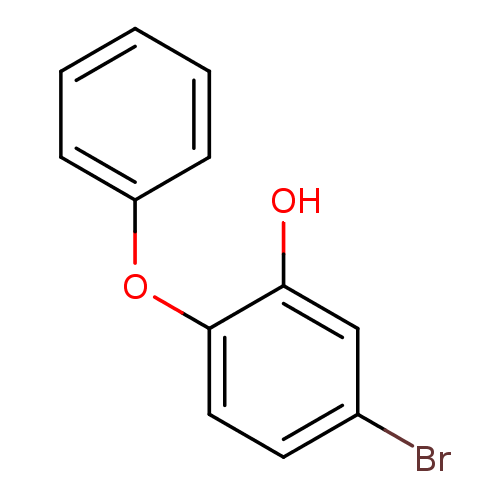
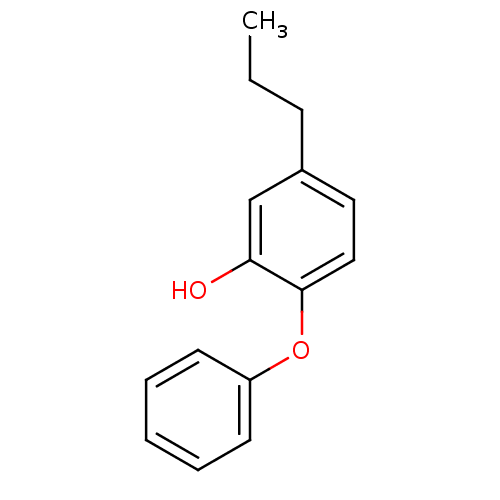
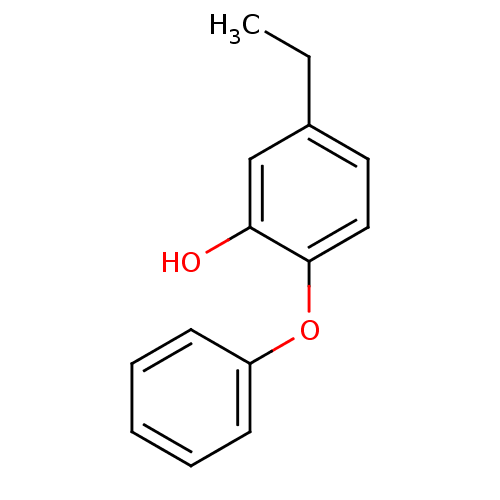
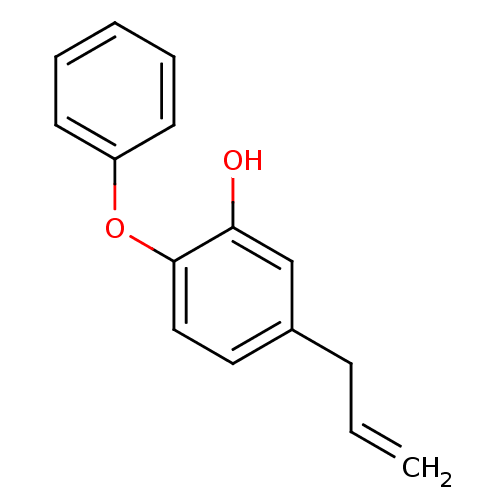
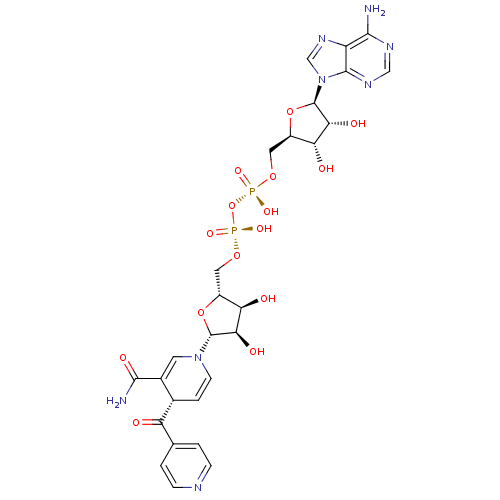
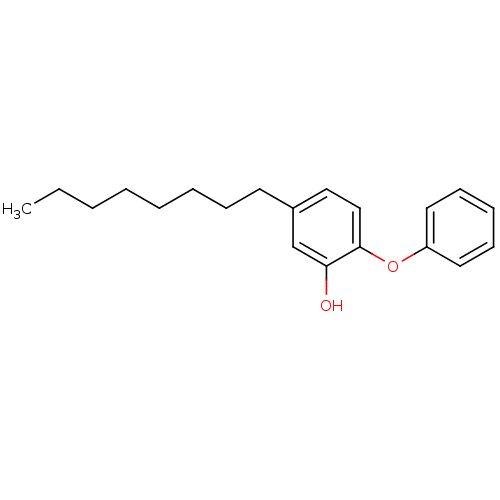
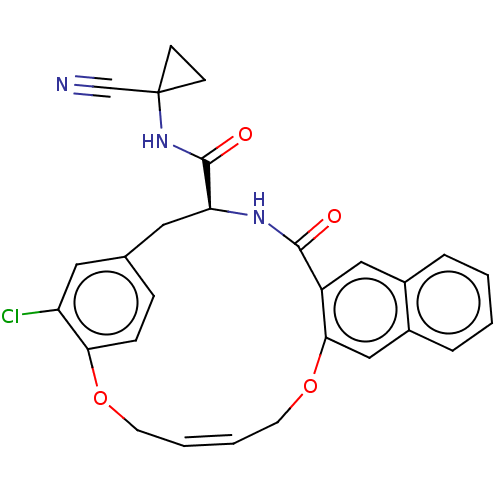
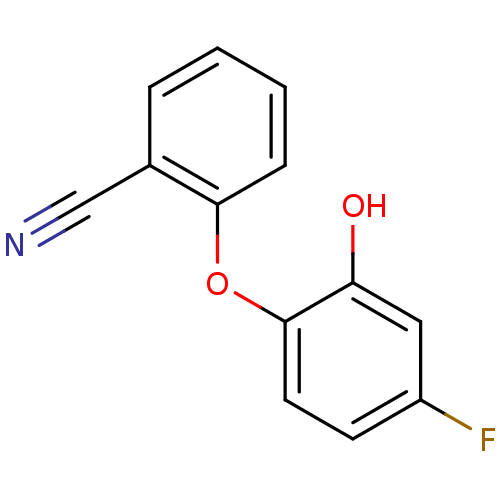
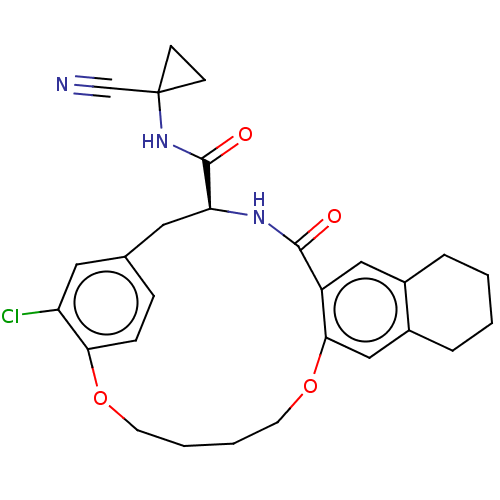
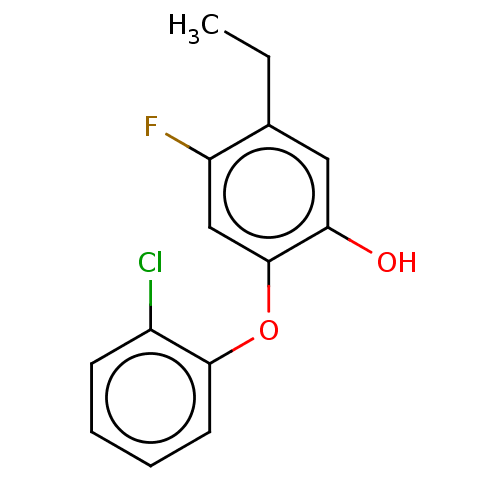
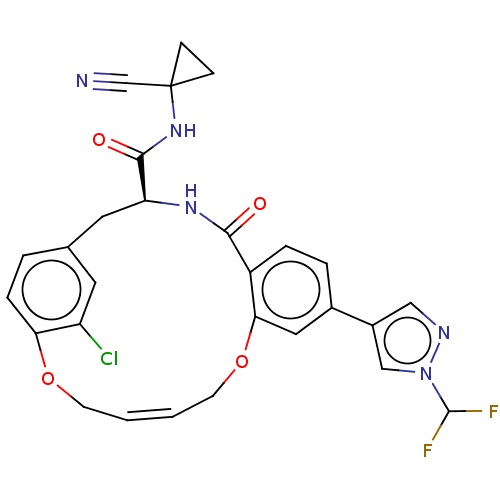
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.0100nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.0100nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.0400nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.0500nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.0500nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
Affinity DataKi: 0.0510nM ΔG°: -58.7kJ/molepH: 8.0 T: 2°CAssay Description:Inhibition constant binding to E-NAD+More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.0600nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.0600nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.0700nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.0900nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.100nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.110nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.120nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.120nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.380nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
Affinity DataKi: 0.440nM ΔG°: -53.4kJ/molepH: 8.0 T: 2°CAssay Description:Inhibition constant binding to E-NAD+More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.5nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Burkholderia pseudomallei)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.510nM ΔG°: -53.0kJ/molepH: 8.0 T: 2°CAssay Description:Slow-onset inhibition kinetics were monitored at 340 nm on a Cary 100 spectrophotometer (Varian) at 25 °C in 30 mM PIPES buffer (pH 8.0) containing 1...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
Suny Stony Brook
Suny Stony Brook
Affinity DataKi: 0.75nM ΔG°: -51.6kJ/molepH: 6.8 T: 2°CAssay Description:Inhibition constants (Ki) were calculated by determining the kcat and Km (DDCoA) values at several fixed inhibitor concentrations. The inhibition dat...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.820nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Burkholderia pseudomallei)
Stony Brook University
Stony Brook University
Affinity DataKi: 1nM ΔG°: -51.4kJ/molepH: 8.0 T: 2°CAssay Description:Slow-onset inhibition kinetics were monitored at 340 nm on a Cary 100 spectrophotometer (Varian) at 25 °C in 30 mM PIPES buffer (pH 8.0) containing 1...More data for this Ligand-Target Pair
Affinity DataKi: 1nMAssay Description:Inhibition of Trypanosoma brucei rhodesiense rhodesain using Cbz-Phe-Arg-AMC as substrate by fluorimetric methodMore data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 1.01nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
Suny Stony Brook
Suny Stony Brook
Affinity DataKi: 1.10nM ΔG°: -50.6kJ/molepH: 6.8 T: 2°CAssay Description:Inhibition constants (Ki) were calculated by determining the kcat and Km (DDCoA) values at several fixed inhibitor concentrations. The inhibition dat...More data for this Ligand-Target Pair
Affinity DataKi: 1.20nMAssay Description:Inhibition of human CatL using Cbz-Phe-Arg-AMC as substrate measured over 30 mins by fluorimetric methodMore data for this Ligand-Target Pair
Affinity DataKi: 1.30nM ΔG°: -50.7kJ/molepH: 8.0 T: 2°CAssay Description:Inhibition constant binding to E-NAD+More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
University Of Wuerzburg
University Of Wuerzburg
Affinity DataKi: 1.30nMpH: 7.5Assay Description:Kinetics were performed on a Cary 100 spectrophotometer (Varian) at 20 °C. Reaction velocities were measured by monitoring the oxidation of NAD(P)H t...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 1.42nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 1.49nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
Affinity DataKi: 1.90nM ΔG°: -49.8kJ/molepH: 8.0 T: 2°CAssay Description:Inhibition constant binding to E-NAD+More data for this Ligand-Target Pair
Affinity DataKi: 1.90nM ΔG°: -49.8kJ/molepH: 8.0 T: 2°CAssay Description:Inhibition constant binding to E-NAD+More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
University Of Wuerzburg
University Of Wuerzburg
Affinity DataKi: 2nMpH: 7.5Assay Description:Kinetics were performed on a Cary 100 spectrophotometer (Varian) at 20 °C. Reaction velocities were measured by monitoring the oxidation of NAD(P)H t...More data for this Ligand-Target Pair
Affinity DataKi: 2nMAssay Description:Inhibition of Trypanosoma brucei rhodesiense rhodesain using Cbz-Phe-Arg-AMC as substrate by fluorimetric methodMore data for this Ligand-Target Pair
Affinity DataKi: 2.10nM ΔG°: -49.5kJ/molepH: 8.0 T: 2°CAssay Description:Inhibition constant binding to E-NAD+More data for this Ligand-Target Pair
Affinity DataKi: 2.30nMAssay Description:Inhibition of human CatL using Cbz-Phe-Arg-AMC as substrate measured over 30 mins by fluorimetric methodMore data for this Ligand-Target Pair
Affinity DataKi: 2.40nMAssay Description:Inhibition of human CatL using Cbz-Phe-Arg-AMC as substrate measured over 30 mins by fluorimetric methodMore data for this Ligand-Target Pair
Affinity DataKi: 2.5nMAssay Description:Inhibition of human CatL using Cbz-Phe-Arg-AMC as substrate measured over 30 mins by fluorimetric methodMore data for this Ligand-Target Pair
Affinity DataKi: 2.5nMAssay Description:Inhibition of Trypanosoma brucei rhodesiense rhodesain using Cbz-Phe-Arg-AMC as substrate by fluorimetric methodMore data for this Ligand-Target Pair
Affinity DataKi: 2.70nM ΔG°: -48.9kJ/molepH: 8.0 T: 2°CAssay Description:Inhibition constant binding to E-NAD+More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
University Of Wuerzburg
University Of Wuerzburg
Affinity DataKi: 2.70nMpH: 7.5Assay Description:Kinetics were performed on a Cary 100 spectrophotometer (Varian) at 20 °C. Reaction velocities were measured by monitoring the oxidation of NAD(P)H t...More data for this Ligand-Target Pair
Affinity DataKi: 3nMAssay Description:Inhibition of human CatL using Cbz-Phe-Arg-AMC as substrate measured over 30 mins by fluorimetric methodMore data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Burkholderia pseudomallei)
Stony Brook University
Stony Brook University
Affinity DataKi: 3nM ΔG°: -48.6kJ/molepH: 8.0 T: 2°CAssay Description:Slow-onset inhibition kinetics were monitored at 340 nm on a Cary 100 spectrophotometer (Varian) at 25 °C in 30 mM PIPES buffer (pH 8.0) containing 1...More data for this Ligand-Target Pair
Affinity DataKi: 3nMAssay Description:Inhibition of Trypanosoma brucei rhodesiense rhodesain using Cbz-Phe-Arg-AMC as substrate by fluorimetric methodMore data for this Ligand-Target Pair
Affinity DataKi: 3.70nMAssay Description:Inhibition of human CatL using Cbz-Phe-Arg-AMC as substrate measured over 30 mins by fluorimetric methodMore data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Burkholderia pseudomallei)
Stony Brook University
Stony Brook University
Affinity DataKi: 4nM ΔG°: -47.9kJ/molepH: 8.0 T: 2°CAssay Description:Slow-onset inhibition kinetics were monitored at 340 nm on a Cary 100 spectrophotometer (Varian) at 25 °C in 30 mM PIPES buffer (pH 8.0) containing 1...More data for this Ligand-Target Pair
Affinity DataKi: 4.60nMAssay Description:Inhibition of human CatL using Cbz-Phe-Arg-AMC as substrate measured over 30 mins by fluorimetric methodMore data for this Ligand-Target Pair
Affinity DataKi: 4.80nMAssay Description:Inhibition of Trypanosoma brucei rhodesiense rhodesain using Cbz-Phe-Arg-AMC as substrate by fluorimetric methodMore data for this Ligand-Target Pair
Affinity DataKi: 4.80nMAssay Description:Agonist activity at human SP/Myc epitope-tagged muscarinic M1 receptor expressed in HEK293T cells assessed as RLuc8-fused Galphaq activation after 2 ...More data for this Ligand-Target Pair
Affinity DataKi: 5.20nMAssay Description:Inhibition of Trypanosoma brucei rhodesiense rhodesain using Cbz-Phe-Arg-AMC as substrate by fluorimetric methodMore data for this Ligand-Target Pair
Affinity DataKi: 5.40nMAssay Description:Inhibition of Trypanosoma brucei rhodesiense rhodesain using Cbz-Phe-Arg-AMC as substrate by fluorimetric methodMore data for this Ligand-Target Pair

 3D Structure (crystal)
3D Structure (crystal)