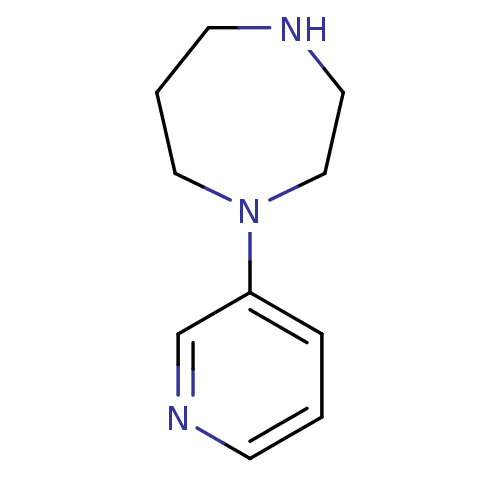
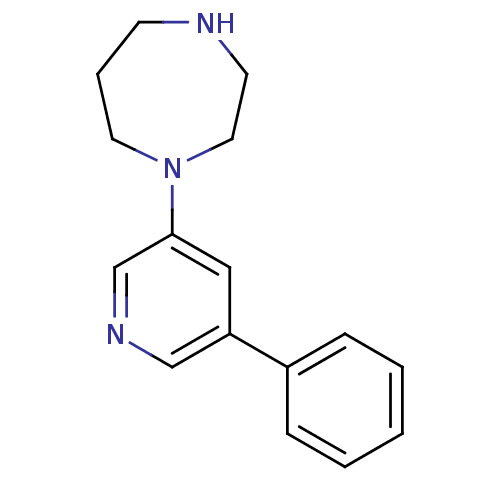
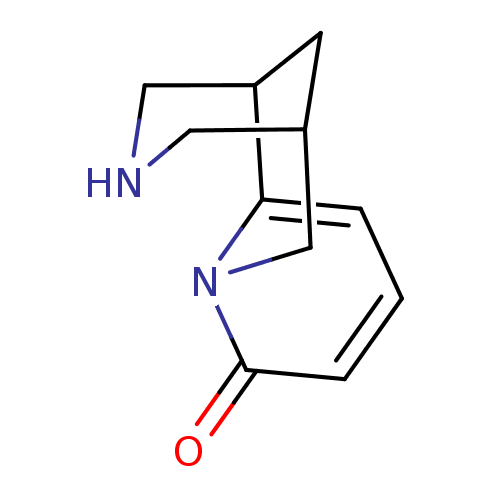
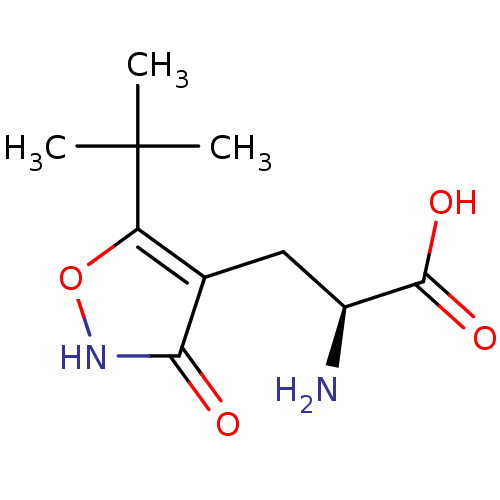
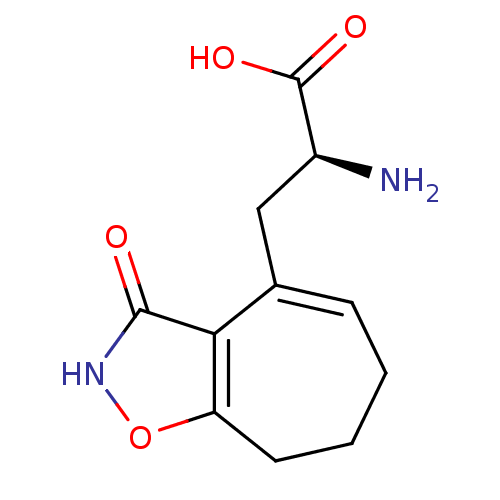
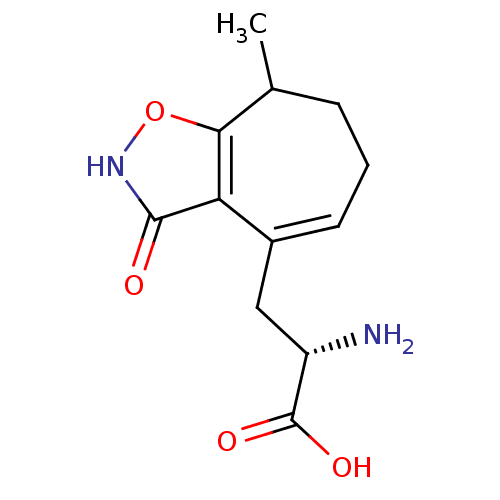
Affinity DataKi: 0.0670nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
Affinity DataKi: 0.0970nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
TargetNeuronal acetylcholine receptor subunit alpha-4(Rattus norvegicus (Rat))
University Of Copenhagen
University Of Copenhagen
Affinity DataKi: 0.25nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
TargetNeuronal acetylcholine receptor subunit alpha-4(Rattus norvegicus (Rat))
University Of Copenhagen
University Of Copenhagen
Affinity DataKi: 0.320nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
TargetNeuronal acetylcholine receptor subunit alpha-4(Rattus norvegicus (Rat))
University Of Copenhagen
University Of Copenhagen
Affinity DataKi: 0.330nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
TargetNeuronal acetylcholine receptor subunit alpha-4(Rattus norvegicus (Rat))
University Of Copenhagen
University Of Copenhagen
Affinity DataKi: 0.620nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
TargetNeuronal acetylcholine receptor subunit alpha-4(Rattus norvegicus (Rat))
University Of Copenhagen
University Of Copenhagen
Affinity DataKi: 0.720nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
TargetNeuronal acetylcholine receptor subunit alpha-4(Rattus norvegicus (Rat))
University Of Copenhagen
University Of Copenhagen
Affinity DataKi: 0.800nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
TargetNeuronal acetylcholine receptor subunit alpha-4(Rattus norvegicus (Rat))
University Of Copenhagen
University Of Copenhagen
Affinity DataKi: 0.950nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
Affinity DataKi: 1.30nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
Affinity DataKi: 2.13nMAssay Description:Displacement of [3H]SYM from rat recombinant GluR5(Q) RNA-edited isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 2.20nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
Affinity DataKi: 2.57nMAssay Description:Displacement of [3H]SYM from rat recombinant GluR5(Q) RNA-edited isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 3.10nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
Affinity DataKi: 5.08nMAssay Description:Displacement of [3H]SYM from rat recombinant GluR5(Q) RNA-edited isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
TargetNeuronal acetylcholine receptor subunit alpha-4(Rattus norvegicus (Rat))
University Of Copenhagen
University Of Copenhagen
Affinity DataKi: 6nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
Affinity DataKi: 8.20nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
Affinity DataKi: 8.90nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
TargetNeuronal acetylcholine receptor subunit alpha-7(Rattus norvegicus (Rat))
University Of Copenhagen
University Of Copenhagen
Affinity DataKi: 18nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
Affinity DataKi: 83nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
TargetNeuronal acetylcholine receptor subunit alpha-7(Rattus norvegicus (Rat))
University Of Copenhagen
University Of Copenhagen
Affinity DataKi: 136nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
TargetNeuronal acetylcholine receptor subunit alpha-7(Rattus norvegicus (Rat))
University Of Copenhagen
University Of Copenhagen
Affinity DataKi: 170nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
Affinity DataKi: 173nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR3 flop isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 175nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR2(R) RNA-edited flop isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
TargetNeuronal acetylcholine receptor subunit alpha-7(Rattus norvegicus (Rat))
University Of Copenhagen
University Of Copenhagen
Affinity DataKi: 190nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, kainate 3(Rattus norvegicus)
University Of Copenhagen
Curated by ChEMBL
University Of Copenhagen
Curated by ChEMBL
Affinity DataKi: 199nMAssay Description:Displacement of [3H]SYM from rat recombinant GluR7A expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 202nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR4 flop isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 232nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR1 flop isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
TargetNeuronal acetylcholine receptor subunit alpha-7(Rattus norvegicus (Rat))
University Of Copenhagen
University Of Copenhagen
Affinity DataKi: 295nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
TargetNeuronal acetylcholine receptor subunit alpha-7(Rattus norvegicus (Rat))
University Of Copenhagen
University Of Copenhagen
Affinity DataKi: 847nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
Affinity DataKi: 1.11E+3nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR3 flop isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, kainate 3(Rattus norvegicus)
University Of Copenhagen
Curated by ChEMBL
University Of Copenhagen
Curated by ChEMBL
Affinity DataKi: 1.38E+3nMAssay Description:Displacement of [3H]SYM from rat recombinant GluR7A expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 1.55E+3nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR1 flop isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 1.66E+3nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR2(R) RNA-edited flop isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 2.04E+3nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR3 flop isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 2.14E+3nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR4 flop isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 2.20E+3nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR4 flop isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, kainate 3(Rattus norvegicus)
University Of Copenhagen
Curated by ChEMBL
University Of Copenhagen
Curated by ChEMBL
Affinity DataKi: 2.53E+3nMAssay Description:Displacement of [3H]SYM from rat recombinant GluR7A expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 2.78E+3nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR1 flop isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 6.31E+3nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR2(R) RNA-edited flop isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
TargetNeuronal acetylcholine receptor subunit alpha-7(Rattus norvegicus (Rat))
University Of Copenhagen
University Of Copenhagen
Affinity DataKi: 9.97E+3nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
TargetNeuronal acetylcholine receptor subunit alpha-7(Rattus norvegicus (Rat))
University Of Copenhagen
University Of Copenhagen
Affinity DataKi: >1.00E+4nMAssay Description:Binding affinities for alpha 4 beta 2 nAChR were determined in tissue suspension of rat cortical membranes using 1 nM [3H]cytisine. Binding affiniti...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, kainate 2(Rattus norvegicus)
University Of Copenhagen
Curated by ChEMBL
University Of Copenhagen
Curated by ChEMBL
Affinity DataKi: >1.00E+5nMAssay Description:Displacement of [3H]KA from rat recombinant GluR6(VCR) RNA-edited isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, kainate 2(Rattus norvegicus)
University Of Copenhagen
Curated by ChEMBL
University Of Copenhagen
Curated by ChEMBL
Affinity DataKi: >1.00E+5nMAssay Description:Displacement of [3H]KA from rat recombinant GluR6(VCR) RNA-edited isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, kainate 2(Rattus norvegicus)
University Of Copenhagen
Curated by ChEMBL
University Of Copenhagen
Curated by ChEMBL
Affinity DataKi: >1.00E+6nMAssay Description:Displacement of [3H]KA from rat recombinant GluR6(VCR) RNA-edited isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataEC50: 6.20E+4nMAssay Description:Agonist activity at ConA-treated rat GluR1 flip isoform expressed in Xenopus laevis oocytes assessed as glutamic acid-induced maximal current by two-...Checked by AuthorMore data for this Ligand-Target Pair
Affinity DataEC50: 660nMAssay Description:Agonist activity at ConA-treated rat GluR5(Q) RNA-edited isoform expressed in Xenopus laevis oocytes assessed as glutamic acid-induced maximal curren...Checked by AuthorMore data for this Ligand-Target Pair
Affinity DataEC50: 130nMAssay Description:Agonist activity at ConA-treated rat GluR5(Q) RNA-edited isoform expressed in Xenopus laevis oocytes assessed as glutamic acid-induced maximal curren...Checked by AuthorMore data for this Ligand-Target Pair
Affinity DataEC50: 6.20E+4nMAssay Description:Agonist activity at ConA-treated rat GluR1 flip isoform expressed in Xenopus laevis oocytes assessed as glutamic acid-induced maximal current by two-...Checked by AuthorMore data for this Ligand-Target Pair
Affinity DataEC50: 4.50E+3nMAssay Description:Agonist activity at ConA-treated rat GluR1 flip isoform expressed in Xenopus laevis oocytes assessed as glutamic acid-induced maximal current by two-...Checked by AuthorMore data for this Ligand-Target Pair

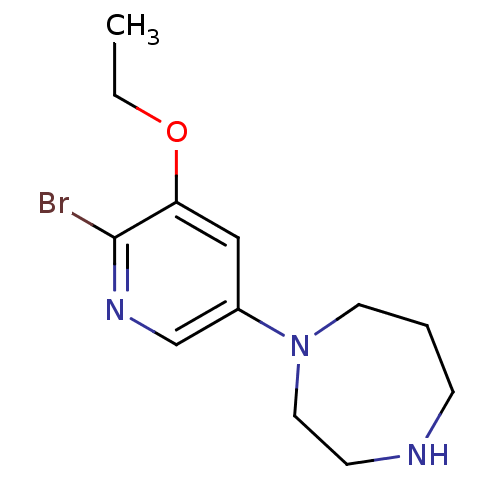
 3D Structure (crystal)
3D Structure (crystal)