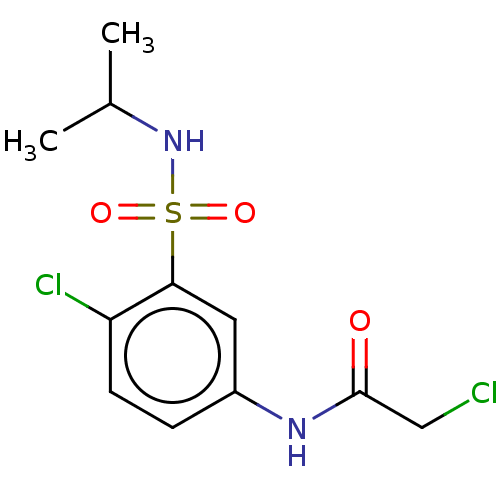
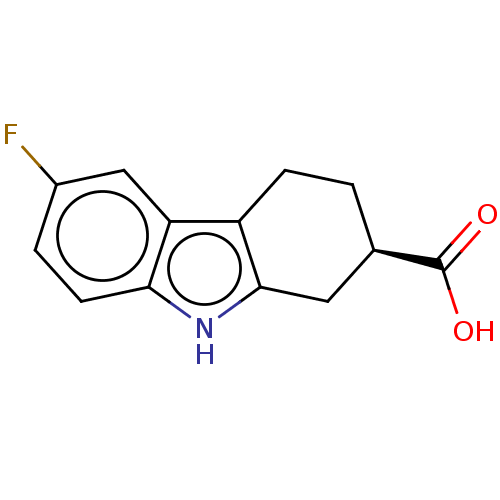
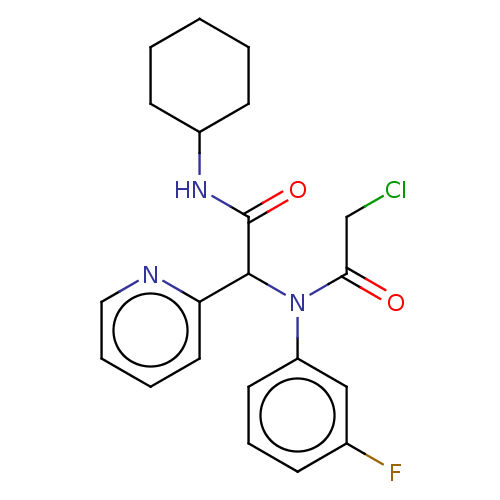
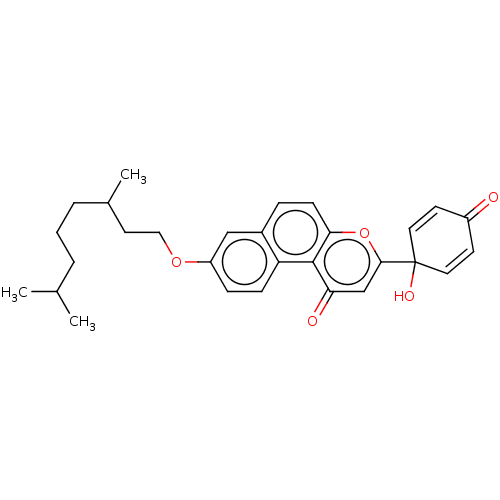
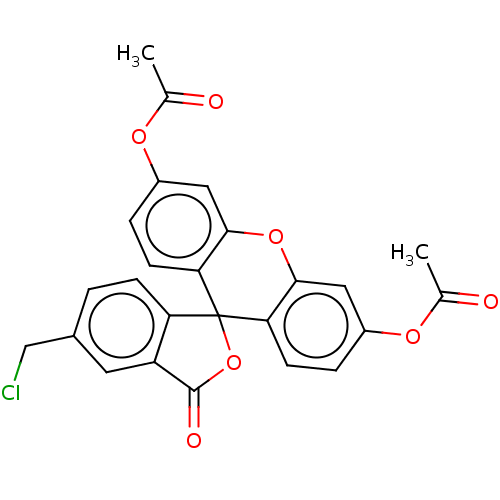
Affinity DataKi: 440nMAssay Description:Inhibition of recombinant GSTO1-1 (unknown origin) expressed in Escherichia coli assessed as inhibitor constant using S-(4-nitrophenacyl)glutathione ...More data for this Ligand-Target Pair
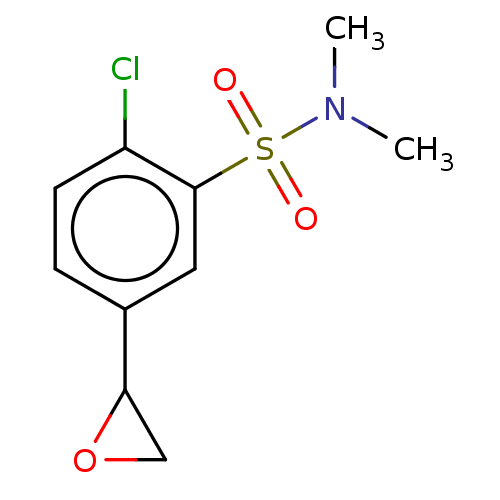
Affinity DataKi: 610nMAssay Description:Inhibition of recombinant GSTO1-1 (unknown origin) expressed in Escherichia coli assessed as inhibitor constant using S-(4-nitrophenacyl)glutathione ...More data for this Ligand-Target Pair
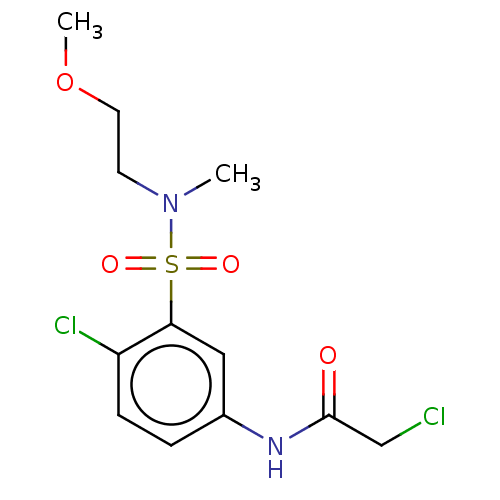
Affinity DataKi: 1.20E+3nMAssay Description:Inhibition of recombinant GSTO1-1 (unknown origin) expressed in Escherichia coli assessed as inhibitor constant using S-(4-nitrophenacyl)glutathione ...More data for this Ligand-Target Pair
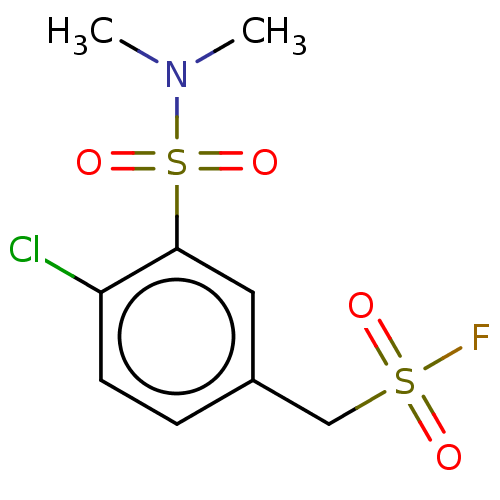
Affinity DataKi: 2.50E+3nMAssay Description:Inhibition of recombinant GSTO1-1 (unknown origin) expressed in Escherichia coli assessed as inhibitor constant using S-(4-nitrophenacyl)glutathione ...More data for this Ligand-Target Pair
Affinity DataKi: 4.80E+3nMAssay Description:Inhibition of recombinant GSTO1-1 (unknown origin) expressed in Escherichia coli assessed as inhibitor constant using S-(4-nitrophenacyl)glutathione ...More data for this Ligand-Target Pair
Affinity DataKi: 5.00E+3nMAssay Description:Inhibition of recombinant GSTO1-1 (unknown origin) expressed in Escherichia coli assessed as inhibitor constant using S-(4-nitrophenacyl)glutathione ...More data for this Ligand-Target Pair
Affinity DataKi: 6.70E+3nMAssay Description:Inhibition of recombinant GSTO1-1 (unknown origin) expressed in Escherichia coli assessed as inhibitor constant using S-(4-nitrophenacyl)glutathione ...More data for this Ligand-Target Pair
Affinity DataKi: 6.90E+3nMAssay Description:Inhibition of recombinant GSTO1-1 (unknown origin) expressed in Escherichia coli assessed as inhibitor constant using S-(4-nitrophenacyl)glutathione ...More data for this Ligand-Target Pair
Affinity DataKi: 9.60E+3nMAssay Description:Inhibition of recombinant GSTO1-1 (unknown origin) expressed in Escherichia coli assessed as inhibitor constant using S-(4-nitrophenacyl)glutathione ...More data for this Ligand-Target Pair
Affinity DataKi: 9.90E+3nMAssay Description:Inhibition of recombinant GSTO1-1 (unknown origin) expressed in Escherichia coli assessed as inhibitor constant using S-(4-nitrophenacyl)glutathione ...More data for this Ligand-Target Pair
TargetBeta sliding clamp(Escherichia coli (strain K12))
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
Affinity DataKi: 1.10E+4nMAssay Description:Inhibition of Escherichia coli DNA sliding clamp protein using N-fluorescein (FAM)-QLDLF-OH tracer by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetBeta sliding clamp(Escherichia coli (strain K12))
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
Affinity DataKi: 1.50E+4nMAssay Description:Inhibition of Escherichia coli DNA sliding clamp protein using N-fluorescein (FAM)-QLDLF-OH tracer by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetBeta sliding clamp(Escherichia coli (strain K12))
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
Affinity DataKi: 1.70E+4nMAssay Description:Inhibition of Escherichia coli DNA sliding clamp protein using N-fluorescein (FAM)-QLDLF-OH tracer by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetBeta sliding clamp(Escherichia coli (strain K12))
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
Affinity DataKi: 2.70E+4nMAssay Description:Inhibition of Escherichia coli DNA sliding clamp protein using N-fluorescein (FAM)-QLDLF-OH tracer by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetBeta sliding clamp(Escherichia coli (strain K12))
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
Affinity DataKi: 3.60E+4nMAssay Description:Inhibition of Escherichia coli DNA sliding clamp protein using N-fluorescein (FAM)-QLDLF-OH tracer by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetBeta sliding clamp(Escherichia coli (strain K12))
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
Affinity DataKi: 4.70E+4nMAssay Description:Inhibition of Escherichia coli DNA sliding clamp protein using N-fluorescein (FAM)-QLDLF-OH tracer by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetBeta sliding clamp(Escherichia coli (strain K12))
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
Affinity DataKi: 6.40E+4nMAssay Description:Inhibition of Escherichia coli DNA sliding clamp protein using N-fluorescein (FAM)-QLDLF-OH tracer by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetBeta sliding clamp(Escherichia coli (strain K12))
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
Affinity DataKi: 7.40E+4nMAssay Description:Inhibition of Escherichia coli DNA sliding clamp protein using N-fluorescein (FAM)-QLDLF-OH tracer by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetBeta sliding clamp(Escherichia coli (strain K12))
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
Affinity DataKi: 8.30E+4nMAssay Description:Inhibition of Escherichia coli DNA sliding clamp protein using N-fluorescein (FAM)-QLDLF-OH tracer by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetBeta sliding clamp(Escherichia coli (strain K12))
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
Affinity DataKi: 8.70E+4nMAssay Description:Inhibition of Escherichia coli DNA sliding clamp protein using N-fluorescein (FAM)-QLDLF-OH tracer by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetBeta sliding clamp(Escherichia coli (strain K12))
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
Affinity DataKi: 9.50E+4nMAssay Description:Inhibition of Escherichia coli DNA sliding clamp protein using N-fluorescein (FAM)-QLDLF-OH tracer by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetBeta sliding clamp(Escherichia coli (strain K12))
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
Affinity DataKi: 9.70E+4nMAssay Description:Inhibition of Escherichia coli DNA sliding clamp protein using N-fluorescein (FAM)-QLDLF-OH tracer by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetBeta sliding clamp(Escherichia coli (strain K12))
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
Affinity DataKi: 1.16E+5nMAssay Description:Inhibition of Escherichia coli DNA sliding clamp protein using N-fluorescein (FAM)-QLDLF-OH tracer by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetBeta sliding clamp(Escherichia coli (strain K12))
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
Affinity DataKi: 1.37E+5nMAssay Description:Inhibition of Escherichia coli DNA sliding clamp protein using N-fluorescein (FAM)-QLDLF-OH tracer by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetBeta sliding clamp(Escherichia coli (strain K12))
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
Affinity DataKi: 1.58E+5nMAssay Description:Inhibition of Escherichia coli DNA sliding clamp protein using N-fluorescein (FAM)-QLDLF-OH tracer by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetBeta sliding clamp(Escherichia coli (strain K12))
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
Affinity DataKi: 1.66E+5nMAssay Description:Inhibition of Escherichia coli DNA sliding clamp protein using N-fluorescein (FAM)-QLDLF-OH tracer by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetBeta sliding clamp(Escherichia coli (strain K12))
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
Affinity DataKi: 1.71E+5nMAssay Description:Inhibition of Escherichia coli DNA sliding clamp protein using N-fluorescein (FAM)-QLDLF-OH tracer by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetBeta sliding clamp(Escherichia coli (strain K12))
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
Affinity DataKi: 1.82E+5nMAssay Description:Inhibition of Escherichia coli DNA sliding clamp protein using N-fluorescein (FAM)-QLDLF-OH tracer by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetBeta sliding clamp(Escherichia coli (strain K12))
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
Affinity DataKi: 1.93E+5nMAssay Description:Inhibition of Escherichia coli DNA sliding clamp protein using N-fluorescein (FAM)-QLDLF-OH tracer by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetBeta sliding clamp(Escherichia coli (strain K12))
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
Affinity DataKi: 2.01E+5nMAssay Description:Inhibition of Escherichia coli DNA sliding clamp protein using N-fluorescein (FAM)-QLDLF-OH tracer by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetBeta sliding clamp(Escherichia coli (strain K12))
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
Affinity DataKi: 2.04E+5nMAssay Description:Inhibition of Escherichia coli DNA sliding clamp protein using N-fluorescein (FAM)-QLDLF-OH tracer by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetBeta sliding clamp(Escherichia coli (strain K12))
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
Affinity DataKi: 2.46E+5nMAssay Description:Inhibition of Escherichia coli DNA sliding clamp protein using N-fluorescein (FAM)-QLDLF-OH tracer by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetBeta sliding clamp(Escherichia coli (strain K12))
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
Affinity DataKi: 4.54E+5nMAssay Description:Inhibition of Escherichia coli DNA sliding clamp protein using N-fluorescein (FAM)-QLDLF-OH tracer by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetBeta sliding clamp(Escherichia coli (strain K12))
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
The University Of Wollongong And The Illawarra Health And Medical Research Institute
Curated by ChEMBL
Affinity DataKi: 4.93E+5nMAssay Description:Inhibition of Escherichia coli DNA sliding clamp protein using N-fluorescein (FAM)-QLDLF-OH tracer by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetA5LHX3/O14818/P20618/P25786/P25787/P25788/P25789/P28062/P28065/P28066/P28070/P28072/P28074/P40306/P49720/P49721/P60900/Q8TAA3/Q99436(Homo sapiens (Human))
Monash University
Curated by ChEMBL
Monash University
Curated by ChEMBL
Affinity DataIC50: <10nMAssay Description:Inhibition of 20S proteasome activity in human ANBL-6 cellsMore data for this Ligand-Target Pair
Affinity DataIC50: 21nMAssay Description:Inhibition of human recombinant GSTO1-1 incubated for 30 mins by fluorescence polarization based gel-based competitive ABPP assayMore data for this Ligand-Target Pair
Affinity DataIC50: 28nMAssay Description:Inhibition of GSTO1-1 (unknown origin) by enzymatic assayMore data for this Ligand-Target Pair
Affinity DataIC50: 30nMAssay Description:Inhibition of recombinant GSTO1-1 (unknown origin) expressed in Escherichia coli using S-(4-nitrophenacyl)glutathione as substrate preincubated for 2...More data for this Ligand-Target Pair
Affinity DataIC50: 31nMAssay Description:Inhibition of GSTO1-1 (unknown origin) by enzymatic assayMore data for this Ligand-Target Pair
Affinity DataIC50: 35nMAssay Description:Inhibition of GSTO1-1 in human MDA-MB-231 cellsMore data for this Ligand-Target Pair
Affinity DataIC50: 40nMAssay Description:Inhibition of GSTO1-1 (unknown origin) by 4-NPG substrate based assayMore data for this Ligand-Target Pair
Affinity DataIC50: 51nMAssay Description:Inhibition of human GSTP1-1 expressed in HEK293 cells by by CDNB-GSH conjugation assayMore data for this Ligand-Target Pair
Affinity DataIC50: 54nMAssay Description:Inhibition of GSTO1-1 (unknown origin) by enzymatic assayMore data for this Ligand-Target Pair
Affinity DataIC50: 60nMAssay Description:Inhibition of recombinant GSTO1-1 (unknown origin) expressed in Escherichia coli using S-(4-nitrophenacyl)glutathione as substrate preincubated for 2...More data for this Ligand-Target Pair
Affinity DataIC50: 70nMAssay Description:Inhibition of recombinant GSTO1-1 (unknown origin) expressed in Escherichia coli using S-(4-nitrophenacyl)glutathione as substrate preincubated for 2...More data for this Ligand-Target Pair
Affinity DataIC50: 80nMAssay Description:Inhibition of recombinant GSTO1-1 (unknown origin) expressed in Escherichia coli using S-(4-nitrophenacyl)glutathione as substrate preincubated for 2...More data for this Ligand-Target Pair
Affinity DataIC50: 110nMAssay Description:Inhibition of recombinant GSTO1-1 (unknown origin) expressed in Escherichia coli using S-(4-nitrophenacyl)glutathione as substrate preincubated for 2...More data for this Ligand-Target Pair
Affinity DataIC50: 120nMAssay Description:Inhibition of GSTO1-1 (unknown origin) by 4-NPG substrate based assayMore data for this Ligand-Target Pair
Affinity DataIC50: 130nMAssay Description:Inhibition of recombinant GSTO1-1 (unknown origin) expressed in Escherichia coli using S-(4-nitrophenacyl)glutathione as substrate preincubated for 2...More data for this Ligand-Target Pair
Affinity DataIC50: 130nMAssay Description:Inhibition of recombinant GSTO1-1 (unknown origin) expressed in Escherichia coli using S-(4-nitrophenacyl)glutathione as substrate preincubated for 2...More data for this Ligand-Target Pair

 3D Structure (crystal)
3D Structure (crystal)