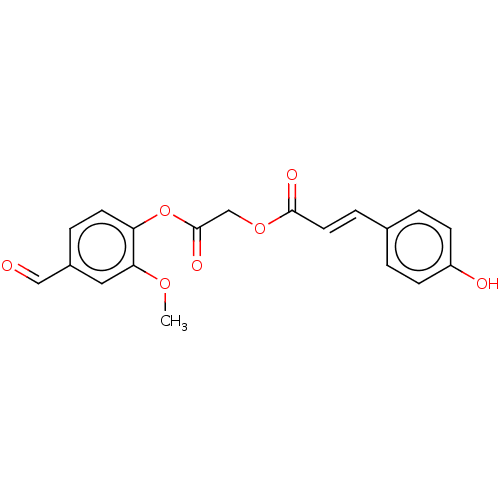
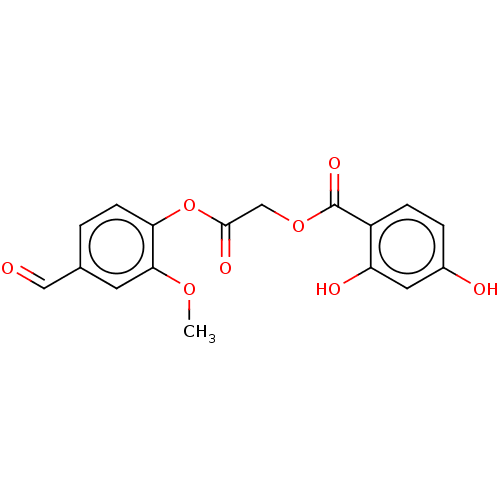
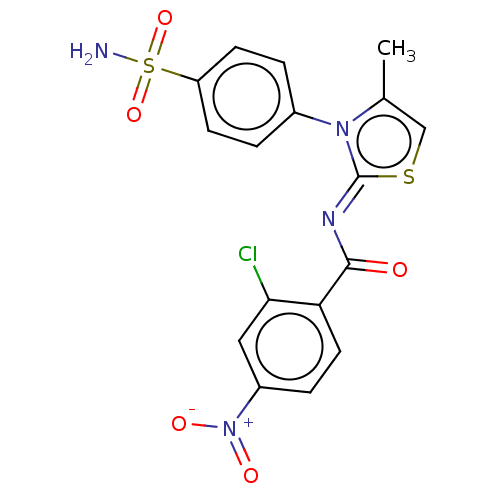
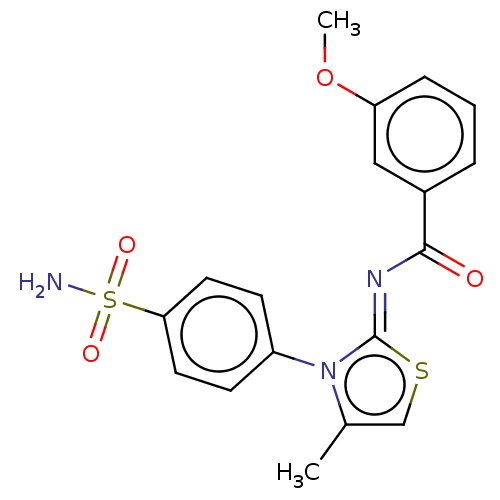
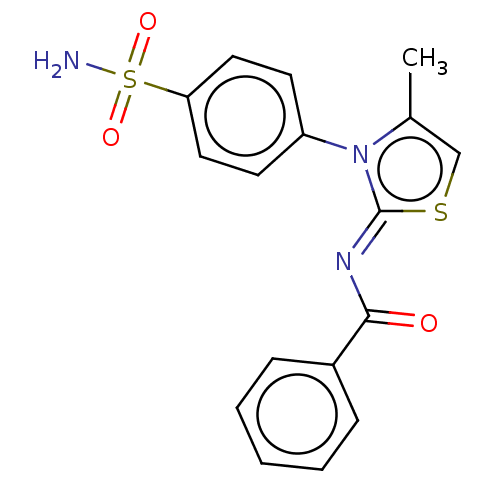
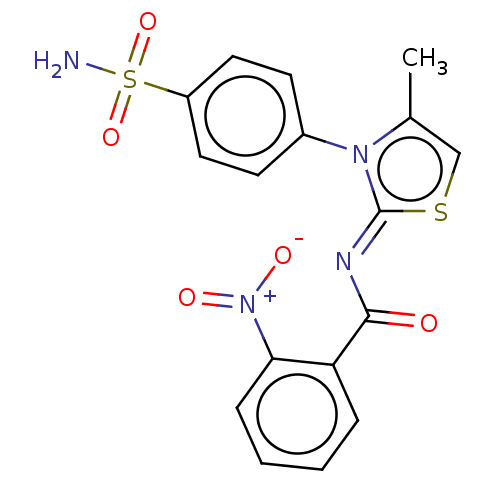
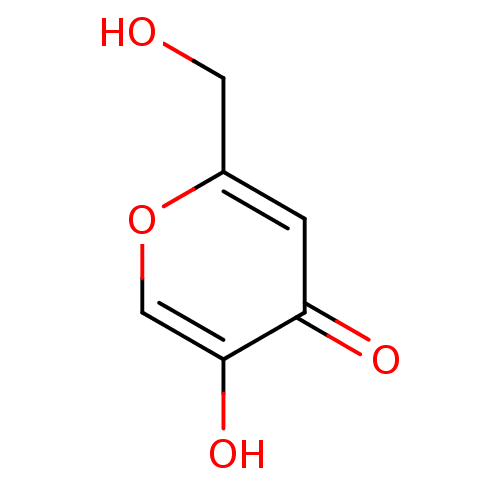
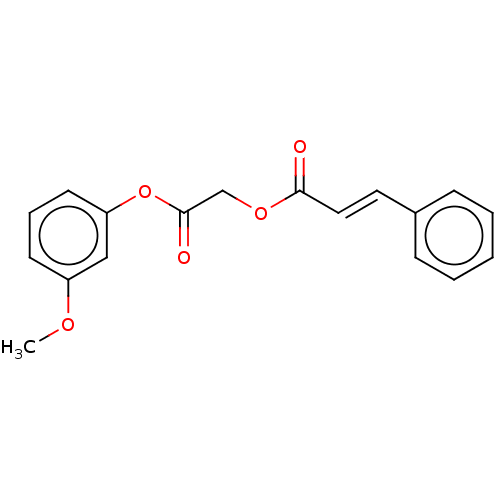
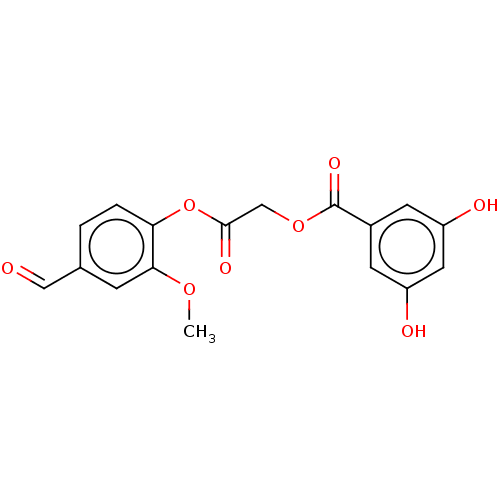
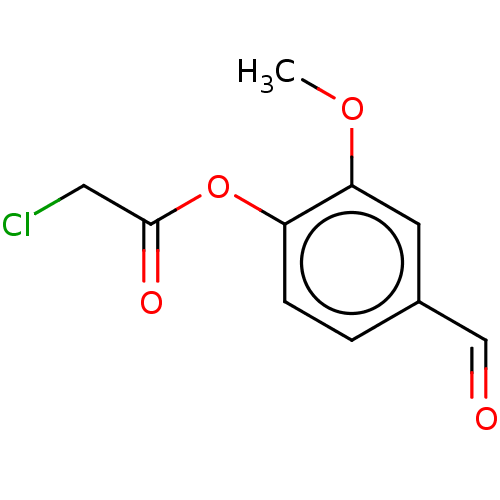
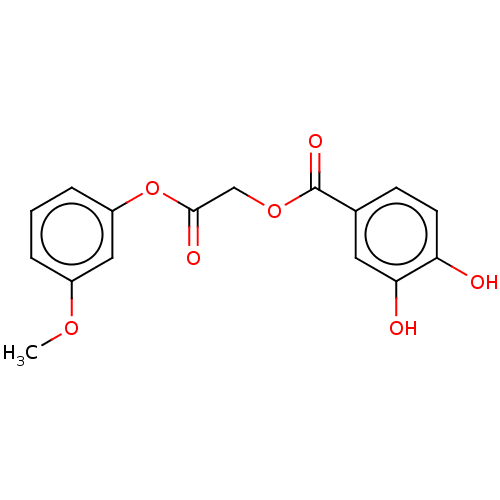
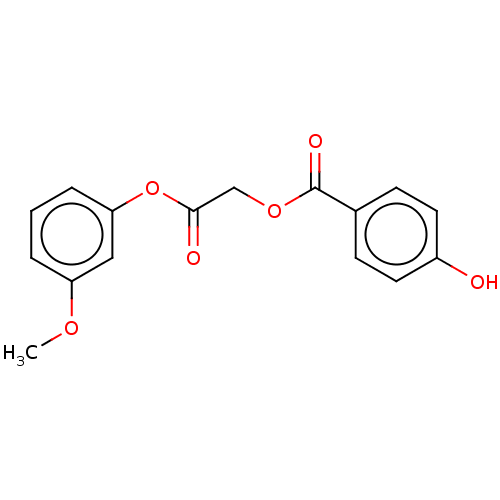
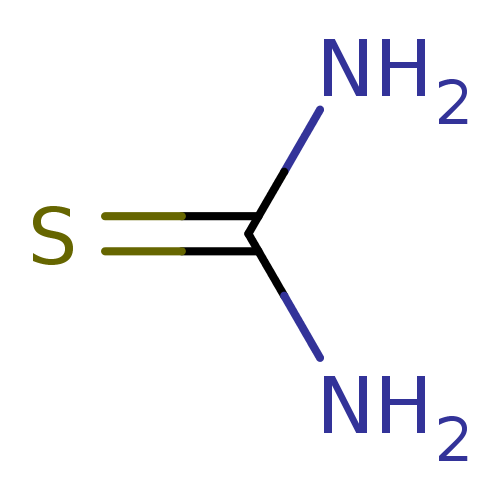TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
The University Of Queensland (Uq)
Curated by ChEMBL
The University Of Queensland (Uq)
Curated by ChEMBL
Affinity DataKi: 1.10E+4nMAssay Description:Non-competitive inhibition of mushroom tyrosinase using L-DOPA as substrate preincubated for 10 mins followed by substrate addition and further incub...More data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
The University Of Queensland (Uq)
Curated by ChEMBL
The University Of Queensland (Uq)
Curated by ChEMBL
Affinity DataKi: 1.30E+4nMAssay Description:Competitive inhibition of mushroom tyrosinase using L-DOPA as substrate assessed as enzyme-inhibitor dissociation constant preincubated for 10 mins f...More data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
The University Of Queensland (Uq)
Curated by ChEMBL
The University Of Queensland (Uq)
Curated by ChEMBL
Affinity DataKi: 4.10E+4nMAssay Description:Competitive inhibition of mushroom tyrosinase using L-DOPA as substrate assessed as enzyme-inhibitor dissociation constant preincubated for 10 mins f...More data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
The University Of Queensland (Uq)
Curated by ChEMBL
The University Of Queensland (Uq)
Curated by ChEMBL
Affinity DataKi: 5.30E+4nMAssay Description:Non-competitive inhibition of mushroom tyrosinase using L-DOPA as substrate assessed as enzyme-substrate-inhibitor dissociation constant preincubated...More data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
The University Of Queensland (Uq)
Curated by ChEMBL
The University Of Queensland (Uq)
Curated by ChEMBL
Affinity DataKi: 6.00E+4nMAssay Description:Competitive inhibition of mushroom tyrosinase using L-DOPA as substrate assessed as enzyme-inhibitor dissociation constant preincubated for 10 mins f...More data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
The University Of Queensland (Uq)
Curated by ChEMBL
The University Of Queensland (Uq)
Curated by ChEMBL
Affinity DataKi: 6.50E+4nMAssay Description:Competitive inhibition of mushroom tyrosinase using L-DOPA as substrate assessed as enzyme-inhibitor dissociation constant preincubated for 10 mins f...More data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
The University Of Queensland (Uq)
Curated by ChEMBL
The University Of Queensland (Uq)
Curated by ChEMBL
Affinity DataKi: 1.00E+5nMAssay Description:Non-competitive inhibition of mushroom tyrosinase using L-DOPA as substrate assessed as enzyme-substrate-inhibitor dissociation constant preincubated...More data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
The University Of Queensland (Uq)
Curated by ChEMBL
The University Of Queensland (Uq)
Curated by ChEMBL
Affinity DataKi: 1.30E+5nMAssay Description:Non-competitive inhibition of mushroom tyrosinase using L-DOPA as substrate preincubated for 10 mins followed by substrate addition and further incub...More data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
The University Of Queensland (Uq)
Curated by ChEMBL
The University Of Queensland (Uq)
Curated by ChEMBL
Affinity DataKi: 2.05E+5nMAssay Description:Non-competitive inhibition of mushroom tyrosinase using L-DOPA as substrate assessed as enzyme-substrate-inhibitor dissociation constant preincubated...More data for this Ligand-Target Pair
Affinity DataIC50: 58nMpH: 8.2Assay Description:The urease inhibitory activity of synthesized sulfonamides (3a-j) was determined by measuring the amount of ammonia produced by the indophenols metho...More data for this Ligand-Target Pair
Affinity DataIC50: 64nMpH: 8.2Assay Description:The urease inhibitory activity of synthesized sulfonamides (3a-j) was determined by measuring the amount of ammonia produced by the indophenols metho...More data for this Ligand-Target Pair
Affinity DataIC50: 72nMpH: 8.2Assay Description:The urease inhibitory activity of synthesized sulfonamides (3a-j) was determined by measuring the amount of ammonia produced by the indophenols metho...More data for this Ligand-Target Pair
Affinity DataIC50: 81nMpH: 8.2Assay Description:The urease inhibitory activity of synthesized sulfonamides (3a-j) was determined by measuring the amount of ammonia produced by the indophenols metho...More data for this Ligand-Target Pair
Affinity DataIC50: 83nMpH: 8.2Assay Description:The urease inhibitory activity of synthesized sulfonamides (3a-j) was determined by measuring the amount of ammonia produced by the indophenols metho...More data for this Ligand-Target Pair
Affinity DataIC50: 83nMpH: 8.2Assay Description:The urease inhibitory activity of synthesized sulfonamides (3a-j) was determined by measuring the amount of ammonia produced by the indophenols metho...More data for this Ligand-Target Pair
Affinity DataIC50: 87nMpH: 8.2Assay Description:The urease inhibitory activity of synthesized sulfonamides (3a-j) was determined by measuring the amount of ammonia produced by the indophenols metho...More data for this Ligand-Target Pair
Affinity DataIC50: 87nMpH: 8.2Assay Description:The urease inhibitory activity of synthesized sulfonamides (3a-j) was determined by measuring the amount of ammonia produced by the indophenols metho...More data for this Ligand-Target Pair
Affinity DataIC50: 96nMpH: 8.2Assay Description:The urease inhibitory activity of synthesized sulfonamides (3a-j) was determined by measuring the amount of ammonia produced by the indophenols metho...More data for this Ligand-Target Pair
Affinity DataIC50: 350nMpH: 8.2Assay Description:The urease inhibitory activity of synthesized sulfonamides (3a-j) was determined by measuring the amount of ammonia produced by the indophenols metho...More data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
The University Of Queensland (Uq)
Curated by ChEMBL
The University Of Queensland (Uq)
Curated by ChEMBL
Affinity DataIC50: 2.30E+3nMAssay Description:Inhibition of mushroom tyrosinase using L-DOPA as substrate by spectrophotometric analysisMore data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
The University Of Queensland (Uq)
Curated by ChEMBL
The University Of Queensland (Uq)
Curated by ChEMBL
Affinity DataIC50: 5.70E+3nMAssay Description:Inhibition of mushroom tyrosinase assessed as reduction in dopachrome formation using L-DOPA as substrate preincubated for 10 mins followed by substr...More data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
The University Of Queensland (Uq)
Curated by ChEMBL
The University Of Queensland (Uq)
Curated by ChEMBL
Affinity DataIC50: 1.61E+4nMAssay Description:Inhibition of mushroom tyrosinase using L-DOPA as substrate preincubated for 10 mins followed by substrate addition measured after 20 minsMore data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
The University Of Queensland (Uq)
Curated by ChEMBL
The University Of Queensland (Uq)
Curated by ChEMBL
Affinity DataIC50: 1.67E+4nMAssay Description:Inhibition of mushroom tyrosinase using L-DOPA as substrate preincubated for 10 mins followed by substrate addition measured after 20 minsMore data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
The University Of Queensland (Uq)
Curated by ChEMBL
The University Of Queensland (Uq)
Curated by ChEMBL
Affinity DataIC50: 1.67E+4nMAssay Description:Inhibition of mushroom tyrosinase assessed as reduction in dopachrome formation using L-DOPA as substrate preincubated for 10 mins followed by substr...More data for this Ligand-Target Pair
Affinity DataIC50: 2.09E+4nMpH: 8.2Assay Description:The urease inhibitory activity of synthesized sulfonamides (3a-j) was determined by measuring the amount of ammonia produced by the indophenols metho...More data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
The University Of Queensland (Uq)
Curated by ChEMBL
The University Of Queensland (Uq)
Curated by ChEMBL
Affinity DataIC50: 2.16E+4nMAssay Description:Inhibition of mushroom tyrosinase using L-DOPA as substrate preincubated for 10 mins followed by substrate addition measured after 20 minsMore data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
The University Of Queensland (Uq)
Curated by ChEMBL
The University Of Queensland (Uq)
Curated by ChEMBL
Affinity DataIC50: 2.38E+4nMAssay Description:Inhibition of mushroom tyrosinase assessed as reduction in dopachrome formation using L-DOPA as substrate preincubated for 10 mins followed by substr...More data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
The University Of Queensland (Uq)
Curated by ChEMBL
The University Of Queensland (Uq)
Curated by ChEMBL
Affinity DataIC50: 3.09E+4nMAssay Description:Inhibition of mushroom tyrosinase using L-DOPA as substrate preincubated for 10 mins followed by substrate addition measured after 20 minsMore data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
The University Of Queensland (Uq)
Curated by ChEMBL
The University Of Queensland (Uq)
Curated by ChEMBL
Affinity DataIC50: 3.94E+4nMAssay Description:Inhibition of mushroom tyrosinase assessed as reduction in dopachrome formation using L-DOPA as substrate preincubated for 10 mins followed by substr...More data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
The University Of Queensland (Uq)
Curated by ChEMBL
The University Of Queensland (Uq)
Curated by ChEMBL
Affinity DataIC50: 4.26E+4nMAssay Description:Inhibition of mushroom tyrosinase using L-DOPA as substrate preincubated for 10 mins followed by substrate addition measured after 20 minsMore data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
The University Of Queensland (Uq)
Curated by ChEMBL
The University Of Queensland (Uq)
Curated by ChEMBL
Affinity DataIC50: 5.98E+4nMAssay Description:Inhibition of mushroom tyrosinase using L-DOPA as substrate preincubated for 10 mins followed by substrate addition measured after 20 minsMore data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
The University Of Queensland (Uq)
Curated by ChEMBL
The University Of Queensland (Uq)
Curated by ChEMBL
Affinity DataIC50: 7.89E+4nMAssay Description:Inhibition of mushroom tyrosinase using L-DOPA as substrate preincubated for 10 mins followed by substrate addition measured after 20 minsMore data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
The University Of Queensland (Uq)
Curated by ChEMBL
The University Of Queensland (Uq)
Curated by ChEMBL
Affinity DataIC50: 1.01E+5nMAssay Description:Inhibition of mushroom tyrosinase using L-DOPA as substrate preincubated for 10 mins followed by substrate addition measured after 20 minsMore data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
The University Of Queensland (Uq)
Curated by ChEMBL
The University Of Queensland (Uq)
Curated by ChEMBL
Affinity DataIC50: 1.24E+5nMAssay Description:Inhibition of mushroom tyrosinase assessed as reduction in dopachrome formation using L-DOPA as substrate preincubated for 10 mins followed by substr...More data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
The University Of Queensland (Uq)
Curated by ChEMBL
The University Of Queensland (Uq)
Curated by ChEMBL
Affinity DataIC50: 1.57E+5nMAssay Description:Inhibition of mushroom tyrosinase using L-DOPA as substrate preincubated for 10 mins followed by substrate addition measured after 20 minsMore data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
The University Of Queensland (Uq)
Curated by ChEMBL
The University Of Queensland (Uq)
Curated by ChEMBL
Affinity DataIC50: 1.73E+5nMAssay Description:Inhibition of mushroom tyrosinase assessed as reduction in dopachrome formation using L-DOPA as substrate preincubated for 10 mins followed by substr...More data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
The University Of Queensland (Uq)
Curated by ChEMBL
The University Of Queensland (Uq)
Curated by ChEMBL
Affinity DataIC50: 1.93E+5nMAssay Description:Inhibition of mushroom tyrosinase assessed as reduction in dopachrome formation using L-DOPA as substrate preincubated for 10 mins followed by substr...More data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
The University Of Queensland (Uq)
Curated by ChEMBL
The University Of Queensland (Uq)
Curated by ChEMBL
Affinity DataIC50: 2.01E+5nMAssay Description:Inhibition of mushroom tyrosinase using L-DOPA as substrate preincubated for 10 mins followed by substrate addition measured after 20 minsMore data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
The University Of Queensland (Uq)
Curated by ChEMBL
The University Of Queensland (Uq)
Curated by ChEMBL
Affinity DataIC50: 2.99E+5nMAssay Description:Inhibition of mushroom tyrosinase assessed as reduction in dopachrome formation using L-DOPA as substrate preincubated for 10 mins followed by substr...More data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
The University Of Queensland (Uq)
Curated by ChEMBL
The University Of Queensland (Uq)
Curated by ChEMBL
Affinity DataIC50: 3.15E+5nMAssay Description:Inhibition of mushroom tyrosinase assessed as reduction in dopachrome formation using L-DOPA as substrate preincubated for 10 mins followed by substr...More data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
The University Of Queensland (Uq)
Curated by ChEMBL
The University Of Queensland (Uq)
Curated by ChEMBL
Affinity DataIC50: 3.38E+5nMAssay Description:Inhibition of mushroom tyrosinase assessed as reduction in dopachrome formation using L-DOPA as substrate preincubated for 10 mins followed by substr...More data for this Ligand-Target Pair