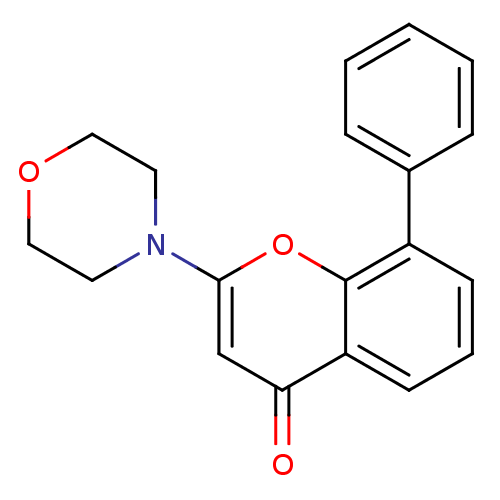
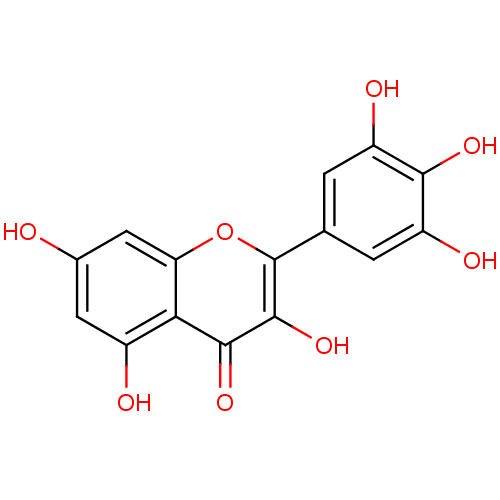
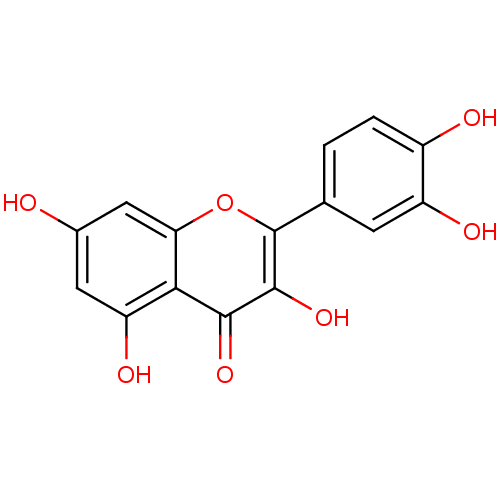
TargetPhosphatidylinositol 3-kinase regulatory subunit alpha/4,5-bisphosphate 3-kinase catalytic subunit alpha isoform(Homo sapiens (Human))
Mrc
Mrc
Affinity DataIC50: 4.20nMpH: 7.4 T: 2°CAssay Description:Ptdlns 3-kinase enzyme activity was assayed by its converting Ptdlns(4,5)P2 into PtdIns(3,4,5)P3 in the presence of radioactive ATP. After reaction,...More data for this Ligand-Target Pair
TargetPhosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform(Homo sapiens (Human))
Mrc
Mrc
Affinity DataIC50: 1.40E+3nM Kd: 210nMpH: 7.2 T: 2°CAssay Description:Binding was detected as a change in the intrinsic tryptophan fluorescence of the PI3K upon the addition of inhibitor. The inhibitor was incubated wit...More data for this Ligand-Target Pair
TargetPhosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform(Homo sapiens (Human))
Mrc
Mrc
Affinity DataIC50: 1.80E+3nM Kd: 170nMpH: 7.2 T: 2°CAssay Description:Binding was detected as a change in the intrinsic tryptophan fluorescence of the PI3K upon the addition of inhibitor. The inhibitor was incubated wit...More data for this Ligand-Target Pair
TargetPhosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform(Homo sapiens (Human))
Mrc
Mrc
Affinity DataIC50: 3.80E+3nM Kd: 280nMpH: 7.2 T: 2°CAssay Description:Binding was detected as a change in the intrinsic tryptophan fluorescence of the PI3K upon the addition of inhibitor. The inhibitor was incubated wit...More data for this Ligand-Target Pair
TargetPhosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform(Homo sapiens (Human))
Mrc
Mrc
Affinity DataIC50: 9.00E+3nM Kd: 290nMpH: 7.2 T: 2°CAssay Description:Binding was detected as a change in the intrinsic tryptophan fluorescence of the PI3K upon the addition of inhibitor. The inhibitor was incubated wit...More data for this Ligand-Target Pair

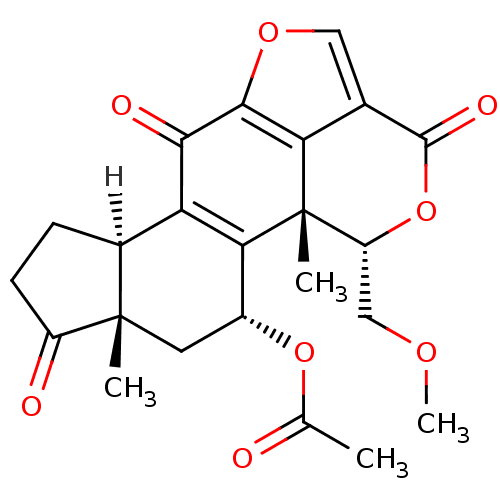
 3D Structure (crystal)
3D Structure (crystal)