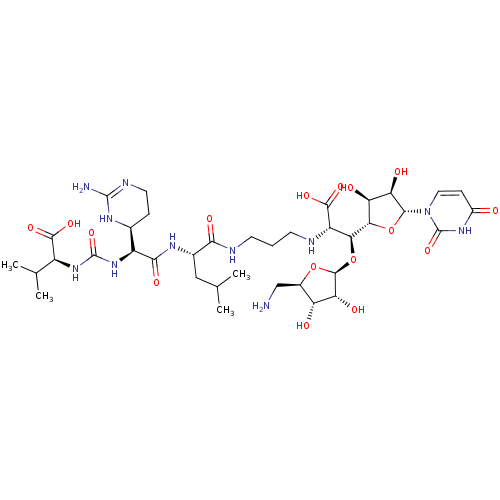
TargetPhospho-N-acetylmuramoyl-pentapeptide-transferase(Bacillus subtilis)
Hokkaido University
Curated by ChEMBL
Hokkaido University
Curated by ChEMBL
Affinity DataKi: 7.60nMAssay Description:Competitive inhibition of Bacillus subtilis MraY using UDP-MurNAc-pentapeptide as substrate after 30 mins by Lineweaver-Burk plotMore data for this Ligand-Target Pair
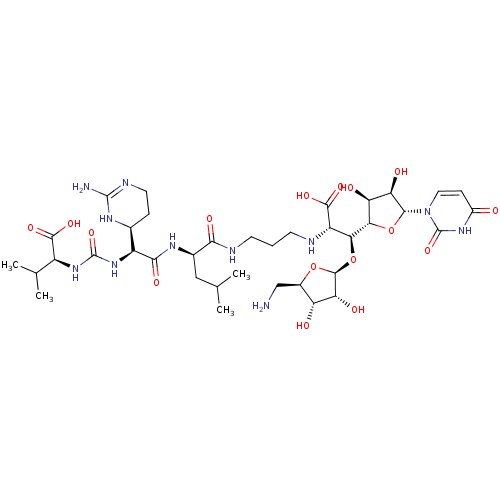
TargetPhospho-N-acetylmuramoyl-pentapeptide-transferase(Bacillus subtilis)
Hokkaido University
Curated by ChEMBL
Hokkaido University
Curated by ChEMBL
Affinity DataKi: 7.60nMAssay Description:Noncompetitive inhibition of Bacillus subtilis MraY using radiolabeled UDP-GlcNAc as substrate after 30 mins by Lineweaver-Burk plotMore data for this Ligand-Target Pair
TargetPhospho-N-acetylmuramoyl-pentapeptide-transferase(Bacillus subtilis)
Hokkaido University
Curated by ChEMBL
Hokkaido University
Curated by ChEMBL
Affinity DataKi: 49nMAssay Description:Competitive inhibition of Bacillus subtilis MraY using UDP-MurNAc-pentapeptide as substrate after 30 mins by Lineweaver-Burk plotMore data for this Ligand-Target Pair
TargetPhospho-N-acetylmuramoyl-pentapeptide-transferase(Bacillus subtilis)
Hokkaido University
Curated by ChEMBL
Hokkaido University
Curated by ChEMBL
Affinity DataKi: 49nMAssay Description:Noncompetitive inhibition of Bacillus subtilis MraY using radiolabeled UDP-GlcNAc as substrate after 30 mins by Lineweaver-Burk plotMore data for this Ligand-Target Pair
TargetPhospho-N-acetylmuramoyl-pentapeptide-transferase(Bacillus subtilis)
Hokkaido University
Curated by ChEMBL
Hokkaido University
Curated by ChEMBL
Affinity DataKi: 247nMAssay Description:Noncompetitive inhibition of Bacillus subtilis MraY using radiolabeled UDP-GlcNAc as substrate after 30 mins by Lineweaver-Burk plotMore data for this Ligand-Target Pair
TargetPhospho-N-acetylmuramoyl-pentapeptide-transferase(Bacillus subtilis)
Hokkaido University
Curated by ChEMBL
Hokkaido University
Curated by ChEMBL
Affinity DataKi: 247nMAssay Description:Competitive inhibition of Bacillus subtilis MraY using UDP-MurNAc-pentapeptide as substrate after 30 mins by Lineweaver-Burk plotMore data for this Ligand-Target Pair
TargetPhospho-N-acetylmuramoyl-pentapeptide-transferase(Bacillus subtilis)
Hokkaido University
Curated by ChEMBL
Hokkaido University
Curated by ChEMBL
Affinity DataKi: 698nMAssay Description:Noncompetitive inhibition of Bacillus subtilis MraY using radiolabeled UDP-GlcNAc as substrate after 30 mins by Lineweaver-Burk plotMore data for this Ligand-Target Pair
TargetPhospho-N-acetylmuramoyl-pentapeptide-transferase(Bacillus subtilis)
Hokkaido University
Curated by ChEMBL
Hokkaido University
Curated by ChEMBL
Affinity DataKi: 698nMAssay Description:Competitive inhibition of Bacillus subtilis MraY using UDP-MurNAc-pentapeptide as substrate after 30 mins by Lineweaver-Burk plotMore data for this Ligand-Target Pair