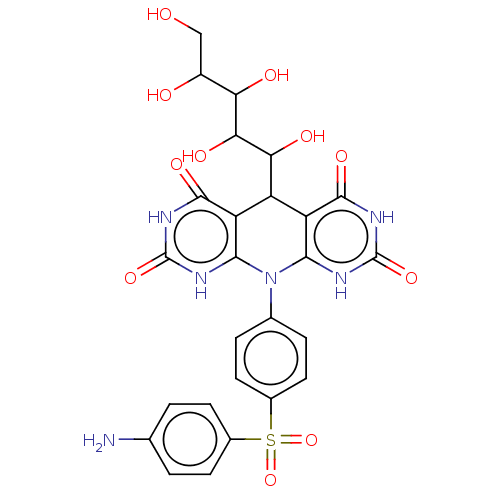
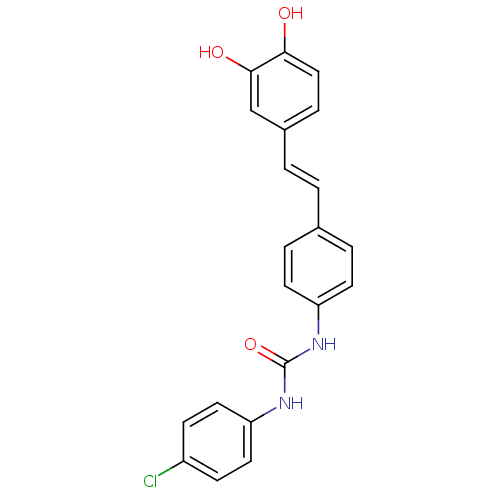
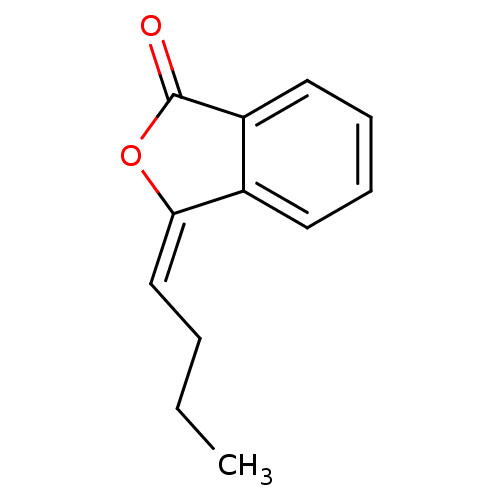
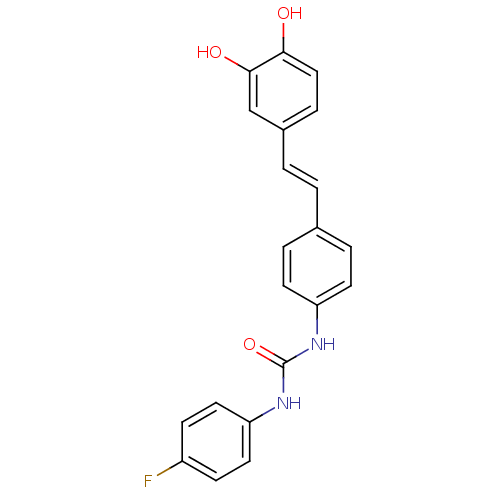
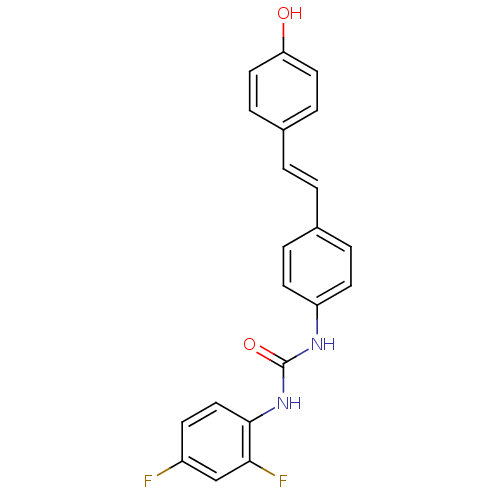
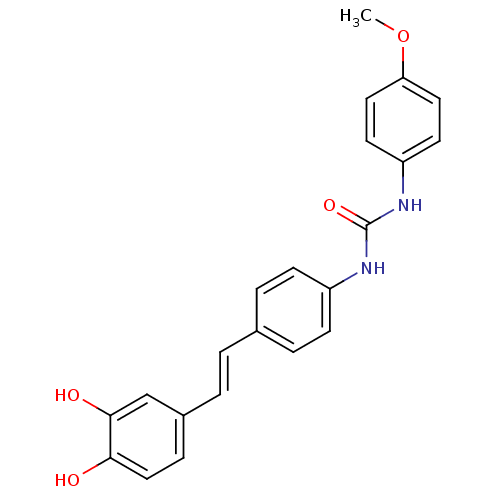
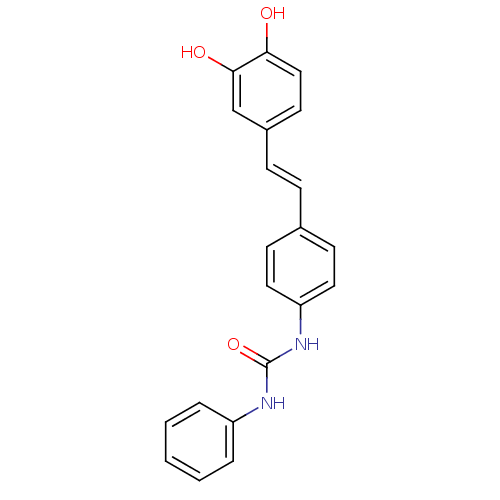
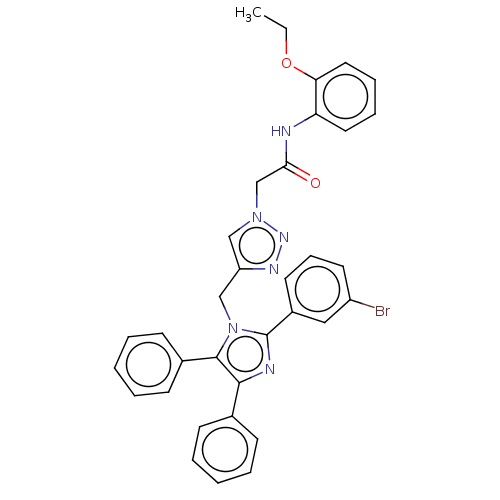
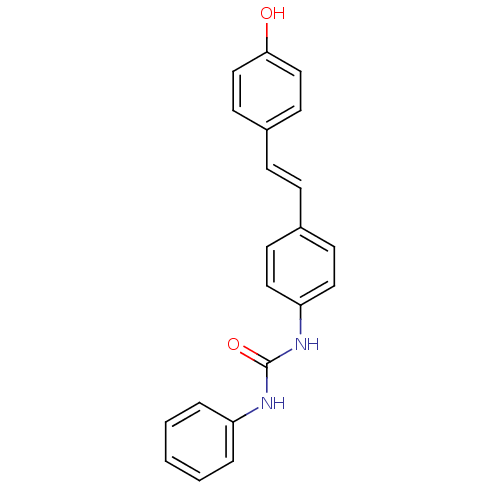
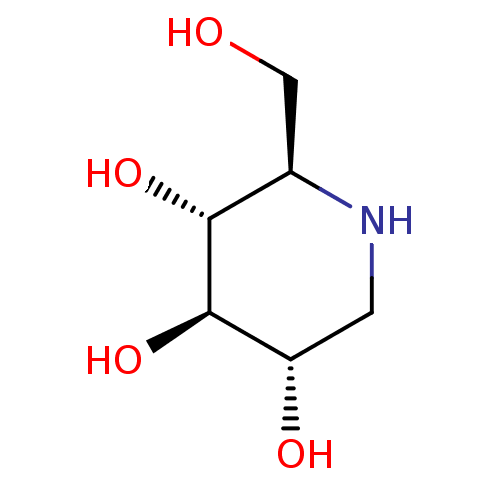
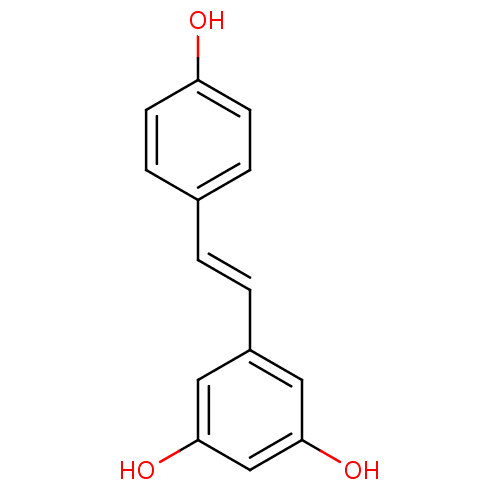
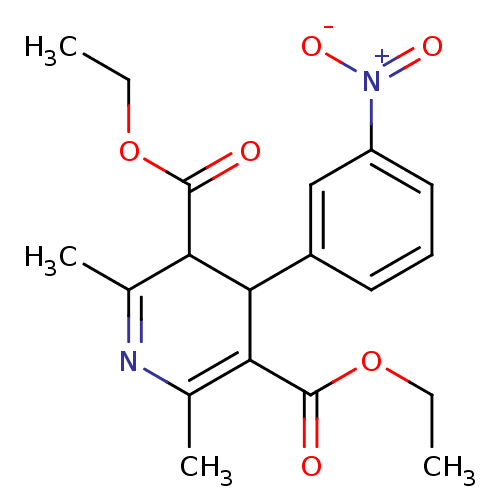
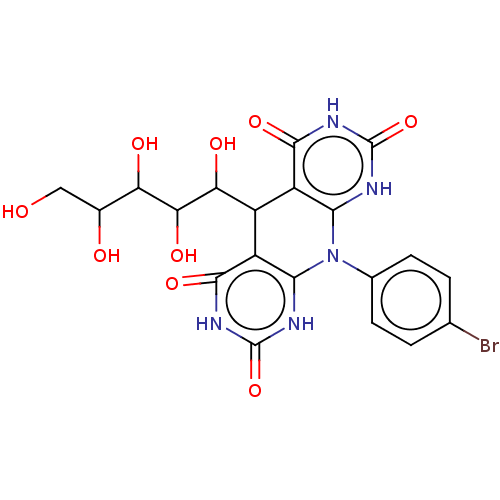
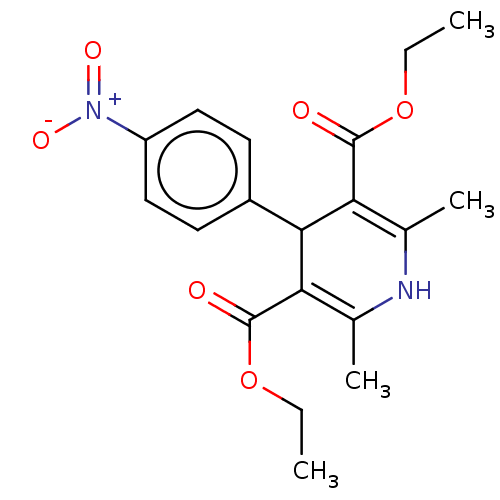
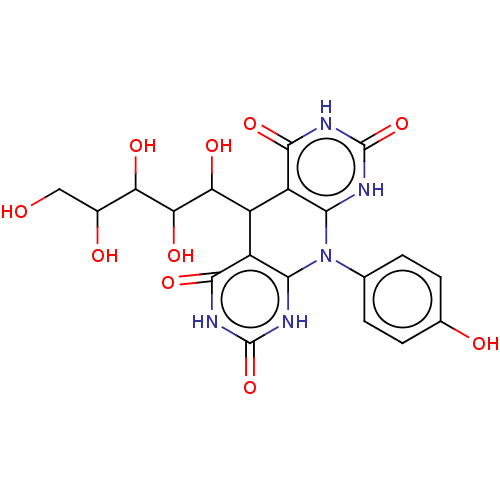
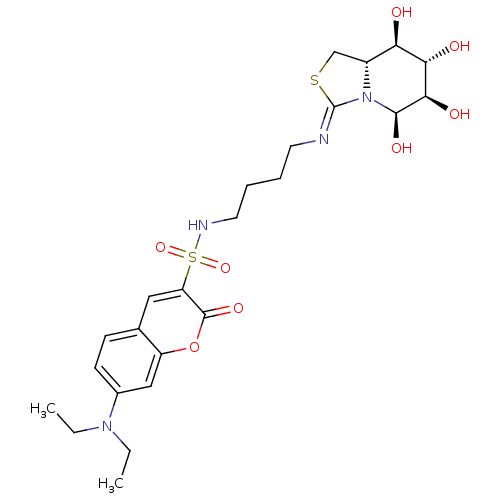
Affinity DataKi: 2.50E+3nMpH: 7.0Assay Description:In this study, the inhibition assay of yeast enzyme was performed in 100 mM phosphate buffer pH 7.0 at 25°C with minor changes, according to the meth...More data for this Ligand-Target Pair
Affinity DataKi: 3.20E+3nM IC50: 8.40E+3nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
Affinity DataKi: 4.60E+3nM IC50: 1.43E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universidad Nacional Aut£Noma De M£Xico
Curated by ChEMBL
Universidad Nacional Aut£Noma De M£Xico
Curated by ChEMBL
Affinity DataKi: 4.86E+3nMAssay Description:Mixed type inhibition of baker's yeast alpha-glucosidase by Dixon plot analysisMore data for this Ligand-Target Pair
Affinity DataKi: 7.20E+3nM IC50: 1.98E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
Affinity DataKi: 9.40E+3nM IC50: 1.85E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
Affinity DataKi: 1.05E+4nM IC50: 1.95E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
Affinity DataKi: 1.06E+4nM IC50: 4.21E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
Affinity DataKi: 1.21E+4nM IC50: 2.88E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
Affinity DataKi: 1.40E+4nMpH: 7.0Assay Description:In this study, the inhibition assay of yeast enzyme was performed in 100 mM phosphate buffer pH 7.0 at 25°C with minor changes, according to the meth...More data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universidad Nacional Aut£Noma De M£Xico
Curated by ChEMBL
Universidad Nacional Aut£Noma De M£Xico
Curated by ChEMBL
Affinity DataKi: 1.48E+4nMAssay Description:Non-competitive inhibition of bakers yeast alpha-glucosidase using p-nitrophenyl-alpha-D-glucopyranoside as substrate by Lineweaver-Burk plot analysi...More data for this Ligand-Target Pair
Affinity DataKi: 1.70E+4nM IC50: 2.91E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
Affinity DataKi: 1.80E+4nM IC50: 3.95E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
Affinity DataKi: 2.15E+4nM IC50: 2.64E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universidad Nacional Aut£Noma De M£Xico
Curated by ChEMBL
Universidad Nacional Aut£Noma De M£Xico
Curated by ChEMBL
Affinity DataKi: 2.50E+4nMAssay Description:Non-competitive inhibition of Saccharomyces cerevisiae alpha-glucosidase using p-nitrophenyl alpha-D-glucopyranoside as substrate preincubated for 15...More data for this Ligand-Target Pair
Affinity DataKi: 3.00E+4nMpH: 7.0Assay Description:In this study, the inhibition assay of yeast enzyme was performed in 100 mM phosphate buffer pH 7.0 at 25°C with minor changes, according to the meth...More data for this Ligand-Target Pair
Affinity DataKi: 3.51E+4nM IC50: 4.64E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universidad Nacional Aut£Noma De M£Xico
Curated by ChEMBL
Universidad Nacional Aut£Noma De M£Xico
Curated by ChEMBL
Affinity DataKi: 6.80E+4nMAssay Description:Competitive inhibition of Saccharomyces cerevisiae alpha-glucosidase using p-nitrophenyl alpha-D-glucopyranoside as substrate preincubated for 15 min...More data for this Ligand-Target Pair
Affinity DataKi: 7.00E+4nMpH: 7.0Assay Description:In this study, the inhibition assay of yeast enzyme was performed in 100 mM phosphate buffer pH 7.0 at 25°C with minor changes, according to the meth...More data for this Ligand-Target Pair
Affinity DataKi: 1.30E+5nMpH: 7.0Assay Description:In this study, the inhibition assay of yeast enzyme was performed in 100 mM phosphate buffer pH 7.0 at 25°C with minor changes, according to the meth...More data for this Ligand-Target Pair
Affinity DataKi: 1.70E+5nMpH: 7.0Assay Description:In this study, the inhibition assay of yeast enzyme was performed in 100 mM phosphate buffer pH 7.0 at 25°C with minor changes, according to the meth...More data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universidad Nacional Aut£Noma De M£Xico
Curated by ChEMBL
Universidad Nacional Aut£Noma De M£Xico
Curated by ChEMBL
Affinity DataKi: 2.46E+5nMAssay Description:Inhibition of yeast alpha-glucosidase after 10 to 30 minsMore data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universidad Nacional Aut£Noma De M£Xico
Curated by ChEMBL
Universidad Nacional Aut£Noma De M£Xico
Curated by ChEMBL
Affinity DataKi: 3.13E+5nMAssay Description:Inhibition of yeast alpha-glucosidase after 10 to 30 minsMore data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universidad Nacional Aut£Noma De M£Xico
Curated by ChEMBL
Universidad Nacional Aut£Noma De M£Xico
Curated by ChEMBL
Affinity DataKi: 4.51E+5nMAssay Description:Inhibition of yeast alpha-glucosidase after 10 to 30 minsMore data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universidad Nacional Aut£Noma De M£Xico
Curated by ChEMBL
Universidad Nacional Aut£Noma De M£Xico
Curated by ChEMBL
Affinity DataKi: 7.14E+5nMAssay Description:Inhibition of yeast alpha-glucosidase after 10 to 30 minsMore data for this Ligand-Target Pair