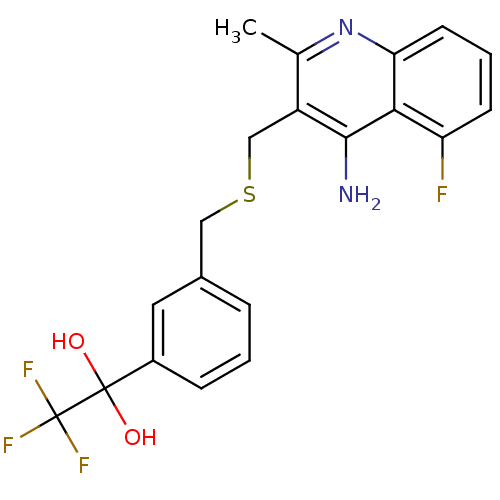
Affinity DataIC50: 1.10E+4nMpH: 7.8 T: 2°CAssay Description:The cholinesterase assays were performed using colorimetric method reported by Ellman. Enzyme activity was determined by measuring the absorbance at ...More data for this Ligand-Target Pair

 3D Structure (crystal)
3D Structure (crystal)