Target2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase(Escherichia coli (strain K12))
University Of Dundee
University Of Dundee
Affinity DataKd: 9.00E+5nMpH: 7.0 T: 2°CAssay Description:An SPR assay was used to determine the binding affinities of compounds. Experiments were performed on a Biacore 3000 (Biacore, Uppsala, Sweden) instr...More data for this Ligand-Target Pair

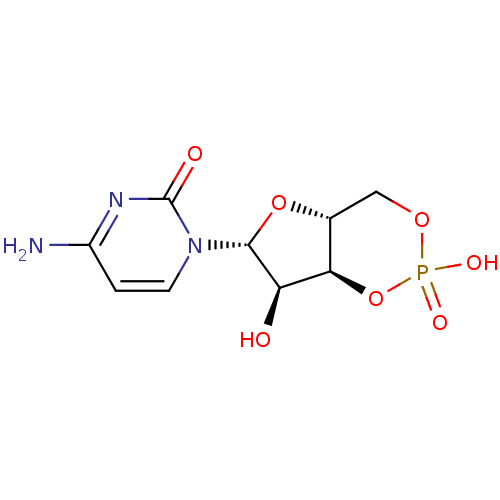
 3D Structure (crystal)
3D Structure (crystal)