TargetBifunctional epoxide hydrolase 2(Homo sapiens (Human))
Boehringer Ingelheim Pharmaceuticals
Curated by ChEMBL
Boehringer Ingelheim Pharmaceuticals
Curated by ChEMBL
Affinity DataIC50: 7nMAssay Description:Inhibition of human soluble EHMore data for this Ligand-Target Pair
TargetBifunctional epoxide hydrolase 2(Homo sapiens (Human))
Boehringer Ingelheim Pharmaceuticals
Curated by ChEMBL
Boehringer Ingelheim Pharmaceuticals
Curated by ChEMBL
Affinity DataIC50: 7nMAssay Description:Displacement of rhodamine-labeled probe from human soluble epoxide hydrolase by fluorescence polarization assayMore data for this Ligand-Target Pair

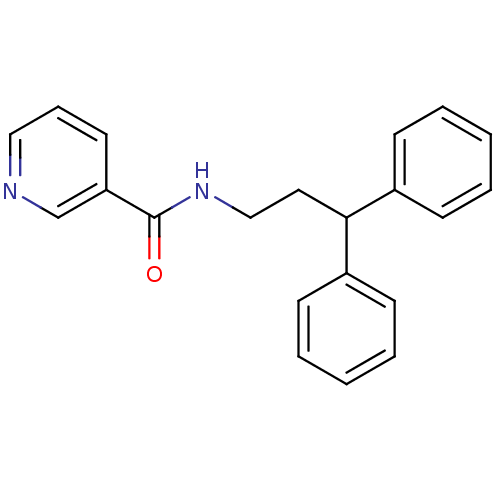
 3D Structure (crystal)
3D Structure (crystal)