TargetBeta-glucuronidase(Escherichia coli (Enterobacteria))
University Of North Carolina At Chapel Hill
University Of North Carolina At Chapel Hill
Affinity DataKi: 960nMAssay Description:Reactions were conducted similarly to the kinetic assays but the reaction consisted of 10 ÁL assay buffer, 5 ÁL inhibitor solution, 5 ÁL of 10 nM enz...More data for this Ligand-Target Pair

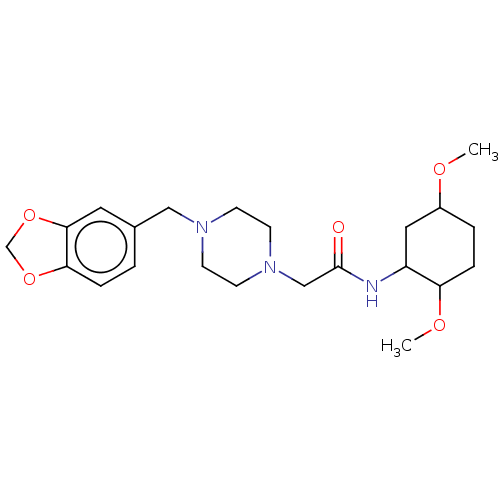
 3D Structure (crystal)
3D Structure (crystal)