TargetGlutamate receptor ionotropic, NMDA 1/2A(Homo sapiens (Human))
Pharmaron-Beijing
Curated by ChEMBL
Pharmaron-Beijing
Curated by ChEMBL
Affinity DataEC50: 382nMAssay Description:Positive allosteric modulation of GluN1/GluN2A NMDAR (unknown origin) expressed in Dox-inducible cells measured every 5 mins by BD calcium indicator ...More data for this Ligand-Target Pair

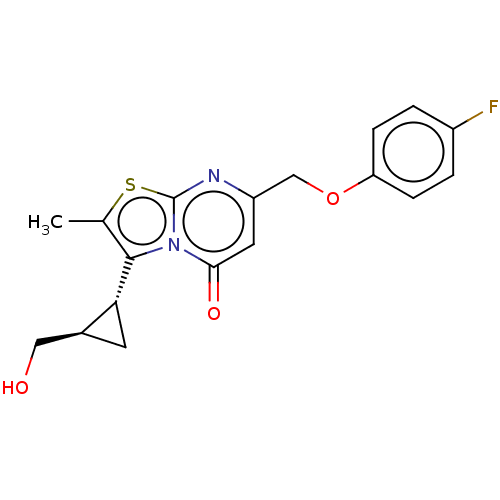
 3D Structure (crystal)
3D Structure (crystal)