Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Coagulation factor VII
Ligand
BDBM12676
Substrate
BDBM12679
Meas. Tech.
Enzyme Assay and Determination of the Inhibition Constants.
pH
7±n/a
Temperature
295.15±n/a K
Ki
>15000±n/a nM
Citation
 Fevig, JM; Cacciola, J; Buriak, J; Rossi, KA; Knabb, RM; Luettgen, JM; Wong, PC; Bai, SA; Wexler, RR; Lam, PY Preparation of 1-(4-methoxyphenyl)-1H-pyrazolo[4,3-d]pyrimidin-7(6H)-ones as potent, selective and bioavailable inhibitors of coagulation factor Xa. Bioorg Med Chem Lett 16:3755-60 (2006) [PubMed] Article
Fevig, JM; Cacciola, J; Buriak, J; Rossi, KA; Knabb, RM; Luettgen, JM; Wong, PC; Bai, SA; Wexler, RR; Lam, PY Preparation of 1-(4-methoxyphenyl)-1H-pyrazolo[4,3-d]pyrimidin-7(6H)-ones as potent, selective and bioavailable inhibitors of coagulation factor Xa. Bioorg Med Chem Lett 16:3755-60 (2006) [PubMed] Article Target
Name:
Coagulation factor VII
Synonyms:
Eptacog alfa | F7 | FA7_HUMAN | Factor VIIa | Factor VIIa (fVIIa) | Proconvertin | SPCA | Thrombin and coagulation factor VII | serum prothrombin conversion accelerator
Type:
Enzyme
Mol. Mass.:
51599.89
Organism:
Homo sapiens (Human)
Description:
n/a
Residue:
466
Sequence:
MVSQALRLLCLLLGLQGCLAAGGVAKASGGETRDMPWKPGPHRVFVTQEEAHGVLHRRRRANAFLEELRPGSLERECKEEQCSFEEAREIFKDAERTKLFWISYSDGDQCASSPCQNGGSCKDQLQSYICFCLPAFEGRNCETHKDDQLICVNENGGCEQYCSDHTGTKRSCRCHEGYSLLADGVSCTPTVEYPCGKIPILEKRNASKPQGRIVGGKVCPKGECPWQVLLLVNGAQLCGGTLINTIWVVSAAHCFDKIKNWRNLIAVLGEHDLSEHDGDEQSRRVAQVIIPSTYVPGTTNHDIALLRLHQPVVLTDHVVPLCLPERTFSERTLAFVRFSLVSGWGQLLDRGATALELMVLNVPRLMTQDCLQQSRKVGDSPNITEYMFCAGYSDGSKDSCKGDSGGPHATHYRGTWYLTGIVSWGQGCATVGHFGVYTRVSQYIEWLQKLMRSEPRPGVLLRAPFP
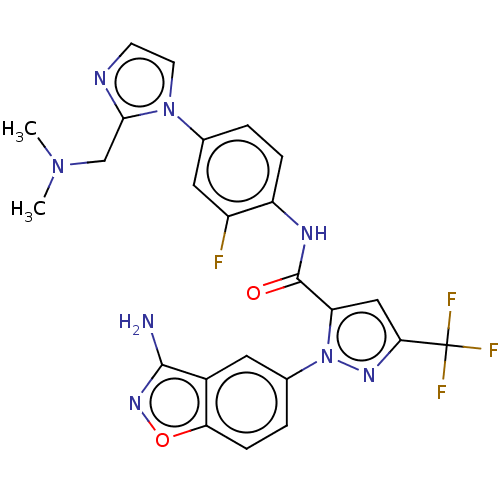
Inhibitor
Name:
BDBM12676
Synonyms:
1-(3-Aminobenzisoxazol-5-yl)-3-trifluoromethyl-N-[2-fluoro-4-[(2-dimethylaminomethyl)imidazol-1-yl]phenyl]-1H-pyrazole-5-carboxyamide Hydrochloride Salt | 1-(3-amino-1,2-benzoxazol-5-yl)-N-(4-{2-[(dimethylamino)methyl]-1H-imidazol-1-yl}-2-fluorophenyl)-3-(trifluoromethyl)-1H-pyrazole-5-carboxamide | BMS-561389 | CHEMBL206335 | CHEMBL558198 | DPC 906 | Razaxaban
Type:
Small organic molecule
Emp. Form.:
C24H20F4N8O2
Mol. Mass.:
528.4616
SMILES:
CN(C)Cc1nccn1-c1ccc(NC(=O)c2cc(nn2-c2ccc3onc(N)c3c2)C(F)(F)F)c(F)c1