Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Tyrosine-protein phosphatase non-receptor type 1
Ligand
BDBM13612
Substrate
BDBM13466
Meas. Tech.
PTP1B Inhibition Assay
pH
7.2±n/a
Temperature
295.15±n/a K
Ki
>100000±n/a nM
Citation
 Bleasdale, JE; Ogg, D; Palazuk, BJ; Jacob, CS; Swanson, ML; Wang, XY; Thompson, DP; Conradi, RA; Mathews, WR; Laborde, AL; Stuchly, CW; Heijbel, A; Bergdahl, K; Bannow, CA; Smith, CW; Svensson, C; Liljebris, C; Schostarez, HJ; May, PD; Stevens, FC; Larsen, SD Small molecule peptidomimetics containing a novel phosphotyrosine bioisostere inhibit protein tyrosine phosphatase 1B and augment insulin action. Biochemistry 40:5642-54 (2001) [PubMed] Article
Bleasdale, JE; Ogg, D; Palazuk, BJ; Jacob, CS; Swanson, ML; Wang, XY; Thompson, DP; Conradi, RA; Mathews, WR; Laborde, AL; Stuchly, CW; Heijbel, A; Bergdahl, K; Bannow, CA; Smith, CW; Svensson, C; Liljebris, C; Schostarez, HJ; May, PD; Stevens, FC; Larsen, SD Small molecule peptidomimetics containing a novel phosphotyrosine bioisostere inhibit protein tyrosine phosphatase 1B and augment insulin action. Biochemistry 40:5642-54 (2001) [PubMed] Article More Info.:
Target
Name:
Tyrosine-protein phosphatase non-receptor type 1
Synonyms:
PTN1_RAT | PTP-1B | PTPase 1B | Protein-Tyrosine Phosphatase 1B (PTP1B) | Ptpn1 | Tyrosine-protein phosphatase, non-receptor type 1
Type:
Enzyme
Mol. Mass.:
49669.91
Organism:
Rattus norvegicus (rat)
Description:
n/a
Residue:
432
Sequence:
MEMEKEFEQIDKAGNWAAIYQDIRHEASDFPCRIAKLPKNKNRNRYRDVSPFDHSRIKLHQEDNDYINASLIKMEEAQRSYILTQGPLPNTCGHFWEMVWEQKSRGVVMLNRIMEKGSLKCAQYWPQKEEKEMVFDDTNLKLTLISEDVKSYYTVRQLELENLATQEAREILHFHYTTWPDFGVPESPASFLNFLFKVRESGSLSPEHGPIVVHCSAGIGRSGTFCLADTCLLLMDKRKDPSSVDIKKVLLEMRRFRMGLIQTADQLRFSYLAVIEGAKFIMGDSSVQDQWKELSHEDLEPPPEHVPPPPRPPKRTLEPHNGKCKELFSNHQWVSEESCEDEDILAREESRAPSIAVHSMSSMSQDTEVRKRMVGGGLQSAQASVPTEEELSPTEEEQKAHRPVHWKPFLVNVCMATALATGAYLCYRVCFH
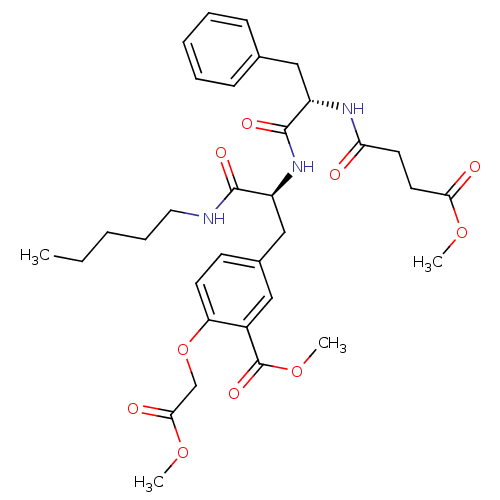
Inhibitor
Name:
BDBM13612
Synonyms:
Compound VII | methyl 2-(2-methoxy-2-oxoethoxy)-5-[(2S)-2-[(2S)-2-(methyl 3-formamidopropanoate)-3-phenylpropanamido]-2-(pentylcarbamoyl)ethyl]benzoate | sulfotyrosyl tripeptide analog 7
Type:
Small organic molecule
Emp. Form.:
C33H43N3O10
Mol. Mass.:
641.7086
SMILES:
CCCCCNC(=O)[C@H](Cc1ccc(OCC(=O)OC)c(c1)C(=O)OC)NC(=O)[C@H](Cc1ccccc1)NC(=O)CCC(=O)OC |r|
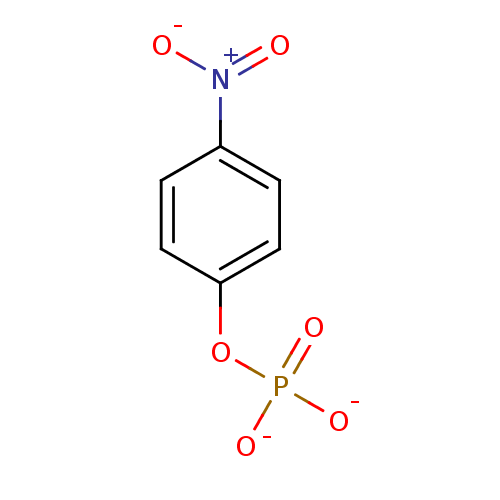
Substrate
Name:
BDBM13466
Synonyms:
4-Nitrophenyl phosphate disodium salt hexahydrate | 4-nitrophenyl phosphate (pNPP) | disodium (4-nitrophenyl) phosphate | para-nitrophenyl phosphate (pNPP)
Type:
Small organic molecule
Emp. Form.:
C6H4NO6P
Mol. Mass.:
217.0739
SMILES:
[O-][N+](=O)c1ccc(O[P+]([O-])([O-])[O-])cc1