Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Corticotropin-releasing factor receptor 1
Ligand
BDBM20974
Substrate
125I-Tyr(0)-ovine-CRF
Meas. Tech.
Receptor Binding Assay
pH
7±n/a
Temperature
296.15±n/a K
IC50
4.2±0.76 nM
Citation
 Dzierba, CD; Tebben, AJ; Wilde, RG; Takvorian, AG; Rafalski, M; Kasireddy-Polam, P; Klaczkiewicz, JD; Pechulis, AD; Davis, AL; Sweet, MP; Woo, AM; Yang, Z; Ebeltoft, SM; Molski, TF; Zhang, G; Zaczek, RC; Trainor, GL; Combs, AP; Gilligan, PJ Dihydropyridopyrazinones and dihydropteridinones as corticotropin-releasing factor-1 receptor antagonists: structure-activity relationships and computational modeling. J Med Chem 50:2269-72 (2007) [PubMed] Article
Dzierba, CD; Tebben, AJ; Wilde, RG; Takvorian, AG; Rafalski, M; Kasireddy-Polam, P; Klaczkiewicz, JD; Pechulis, AD; Davis, AL; Sweet, MP; Woo, AM; Yang, Z; Ebeltoft, SM; Molski, TF; Zhang, G; Zaczek, RC; Trainor, GL; Combs, AP; Gilligan, PJ Dihydropyridopyrazinones and dihydropteridinones as corticotropin-releasing factor-1 receptor antagonists: structure-activity relationships and computational modeling. J Med Chem 50:2269-72 (2007) [PubMed] Article More Info.:
Target
Name:
Corticotropin-releasing factor receptor 1
Synonyms:
CRF-R | CRF1 | CRFR1_RAT | CRH-R 1 | Corticotropin releasing factor receptor | Corticotropin releasing factor receptor 1 | Corticotropin-releasing Factor Receptor 1 | Corticotropin-releasing hormone receptor 1 | Crhr | Crhr1
Type:
G Protein-Coupled Receptor (GPCR)
Mol. Mass.:
47870.75
Organism:
Rattus norvegicus (rat)
Description:
Receptor binding assays were performed using rat cortex homogenate.
Residue:
415
Sequence:
MGRRPQLRLVKALLLLGLNPVSTSLQDQRCENLSLTSNVSGLQCNASVDLIGTCWPRSPAGQLVVRPCPAFFYGVRYNTTNNGYRECLANGSWAARVNYSECQEILNEEKKSKVHYHVAVIINYLGHCISLVALLVAFVLFLRLRSIRCLRNIIHWNLISAFILRNATWFVVQLTVSPEVHQSNVAWCRLVTAAYNYFHVTNFFWMFGEGCYLHTAIVLTYSTDRLRKWMFVCIGWGVPFPIIVAWAIGKLHYDNEKCWFGKRPGVYTDYIYQGPMILVLLINFIFLFNIVRILMTKLRASTTSETIQYRKAVKATLVLLPLLGITYMLFFVNPGEDEVSRVVFIYFNSFLESFQGFFVSVFYCFLNSEVRSAIRKRWRRWQDKHSIRARVARAMSIPTSPTRVSFHSIKQSTAV
Inhibitor
Name:
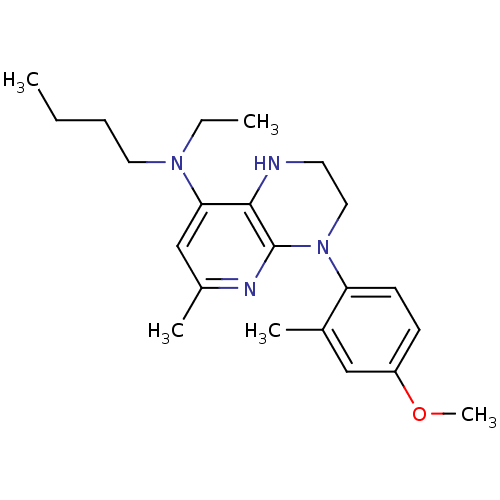
BDBM20974
Synonyms:
N-butyl-N-ethyl-4-(4-methoxy-2-methylphenyl)-6-methyl-1H,2H,3H,4H-pyrido[2,3-b]pyrazin-8-amine | Pyridopyrazine, 1b
Type:
Small organic molecule
Emp. Form.:
C22H32N4O
Mol. Mass.:
368.5157
SMILES:
CCCCN(CC)c1cc(C)nc2N(CCNc12)c1ccc(OC)cc1C
Substrate
Name:
125I-Tyr(0)-ovine-CRF
Synonyms:
Ovine Tyr-CRF | oCRF
Type:
radiolabeled substrate
Mol. Mass.:
4833.46
Organism:
Ovis aries (sheep)
Description:
n/a
Residue:
42
Sequence:
YSQEPPISLDLTFHLLREVLEMTKADQLAQQAHSNRKLLDIA