Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Peptidyl-prolyl cis-trans isomerase FKBP1A
Ligand
BDBM23353
Substrate
BDBM23318
Meas. Tech.
Peptidyl Prolyl Isomerase (PPIase) Inibition Assay
pH
8±n/a
Temperature
288.15±n/a K
Ki
123±n/a nM
Citation
 Hudack, RA; Barta, NS; Guo, C; Deal, J; Dong, L; Fay, LK; Caprathe, B; Chatterjee, A; Vanderpool, D; Bigge, C; Showalter, R; Bender, S; Augelli-Szafran, CE; Lunney, E; Hou, X Design, synthesis, and biological activity of novel polycyclic aza-amide FKBP12 ligands. J Med Chem 49:1202-6 (2006) [PubMed] Article
Hudack, RA; Barta, NS; Guo, C; Deal, J; Dong, L; Fay, LK; Caprathe, B; Chatterjee, A; Vanderpool, D; Bigge, C; Showalter, R; Bender, S; Augelli-Szafran, CE; Lunney, E; Hou, X Design, synthesis, and biological activity of novel polycyclic aza-amide FKBP12 ligands. J Med Chem 49:1202-6 (2006) [PubMed] Article More Info.:
Target
Name:
Peptidyl-prolyl cis-trans isomerase FKBP1A
Synonyms:
12 kDa FK506-binding protein | 12 kDa FKBP | FK506-binding protein 1A | FK506-binding protein 1A (FKBP12) | FKB1A_HUMAN | FKBP-12 | FKBP-1A | FKBP1 | FKBP12 | FKBP1A | Immunophilin FKBP12 | PPIase | PPIase FKBP1A | Peptidyl-prolyl cis-trans isomerase (FKBP) | Rotamase | RyR1/FKBP12 | mTOR/FKBP12A/FKBP12B
Type:
Isomerase
Mol. Mass.:
11953.09
Organism:
Homo sapiens (Human)
Description:
P62942
Residue:
108
Sequence:
MGVQVETISPGDGRTFPKRGQTCVVHYTGMLEDGKKFDSSRDRNKPFKFMLGKQEVIRGWEEGVAQMSVGQRAKLTISPDYAYGATGHPGIIPPHATLVFDVELLKLE
Inhibitor
Name:
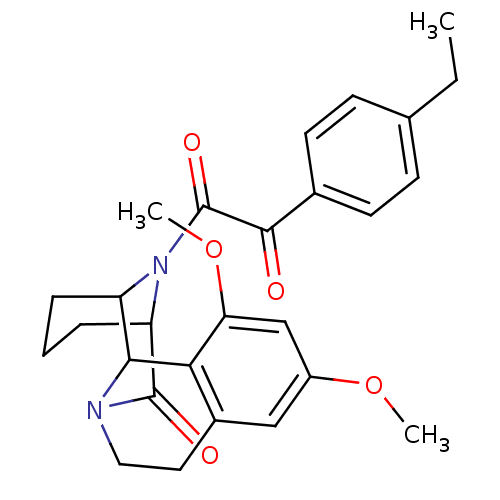
BDBM23353
Synonyms:
1-{4,6-dimethoxy-12-oxo-11,17-diazatetracyclo[11.3.1.0^{2,11}.0^{3,8}]heptadeca-3(8),4,6-trien-17-yl}-2-(4-ethylphenyl)ethane-1,2-dione | Tetracyclic aza-amide, 6i
Type:
Small organic molecule
Emp. Form.:
C27H30N2O5
Mol. Mass.:
462.5375
SMILES:
CCc1ccc(cc1)C(=O)C(=O)N1C2CCCC1C(=O)N1CCc3cc(OC)cc(OC)c3C21 |TLB:10:12:14.15.16:20.33.18|
Substrate
Name:
BDBM23318
Synonyms:
3-{[(1S)-1-{[(2S)-4-methyl-1-[(2S)-2-{[(1S)-1-[(4-nitrophenyl)carbamoyl]-2-phenylethyl]carbamoyl}pyrrolidin-1-yl]-1-oxopentan-2-yl]carbamoyl}ethyl]carbamoyl}propanoic acid | N-Succinyl-L-alanyl-L-Leucyl-L-prolyl-L-phenylalanine 4-nitroanilide | N-succinyl Ala-Leu-Pro-Phe p-nitroanilide | Suc-ALPF-pNA
Type:
Colorimetric substrate
Emp. Form.:
C33H42N6O9
Mol. Mass.:
666.7214
SMILES:
CC(C)C[C@H](NC(=O)[C@H](C)NC(=O)CCC(O)=O)C(=O)N1CCC[C@H]1C(=O)N[C@@H](Cc1ccccc1)C(=O)Nc1ccc(cc1)[N+]([O-])=O |r|