Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Peroxisome proliferator-activated receptor alpha
Ligand
BDBM29895
Substrate
BDBM28662
Meas. Tech.
Scintillation Proximity Assay (Ki) and Cell-Based Transcription Assay (EC50)
pH
7.1±n/a
Temperature
295.15±n/a K
Ki
4100±n/a nM
EC50
>100000±n/a nM
Citation
 Connors, RV; Wang, Z; Harrison, M; Zhang, A; Wanska, M; Hiscock, S; Fox, B; Dore, M; Labelle, M; Sudom, A; Johnstone, S; Liu, J; Walker, NP; Chai, A; Siegler, K; Li, Y; Coward, P Identification of a PPARdelta agonist with partial agonistic activity on PPARgamma. Bioorg Med Chem Lett 19:3550-4 (2009) [PubMed] Article
Connors, RV; Wang, Z; Harrison, M; Zhang, A; Wanska, M; Hiscock, S; Fox, B; Dore, M; Labelle, M; Sudom, A; Johnstone, S; Liu, J; Walker, NP; Chai, A; Siegler, K; Li, Y; Coward, P Identification of a PPARdelta agonist with partial agonistic activity on PPARgamma. Bioorg Med Chem Lett 19:3550-4 (2009) [PubMed] Article Target
Name:
Peroxisome proliferator-activated receptor alpha
Synonyms:
NR1C1 | Nuclear receptor subfamily 1 group C member 1 | PPAR | PPAR alpha/gamma | PPAR-alpha | PPARA | PPARA_HUMAN | Peroxisome Proliferator-Activated Receptor alpha | Peroxisome proliferator-activated receptor | Peroxisome proliferator-activated receptor alpha (PPAR alpha)
Type:
Enzyme
Mol. Mass.:
52222.08
Organism:
Homo sapiens (Human)
Description:
Q07869
Residue:
468
Sequence:
MVDTESPLCPLSPLEAGDLESPLSEEFLQEMGNIQEISQSIGEDSSGSFGFTEYQYLGSCPGSDGSVITDTLSPASSPSSVTYPVVPGSVDESPSGALNIECRICGDKASGYHYGVHACEGCKGFFRRTIRLKLVYDKCDRSCKIQKKNRNKCQYCRFHKCLSVGMSHNAIRFGRMPRSEKAKLKAEILTCEHDIEDSETADLKSLAKRIYEAYLKNFNMNKVKARVILSGKASNNPPFVIHDMETLCMAEKTLVAKLVANGIQNKEAEVRIFHCCQCTSVETVTELTEFAKAIPGFANLDLNDQVTLLKYGVYEAIFAMLSSVMNKDGMLVAYGNGFITREFLKSLRKPFCDIMEPKFDFAMKFNALELDDSDISLFVAAIICCGDRPGLLNVGHIEKMQEGIVHVLRLHLQSNHPDDIFLFPKLLQKMADLRQLVTEHAQLVQIIKKTESDAALHPLLQEIYRDMY
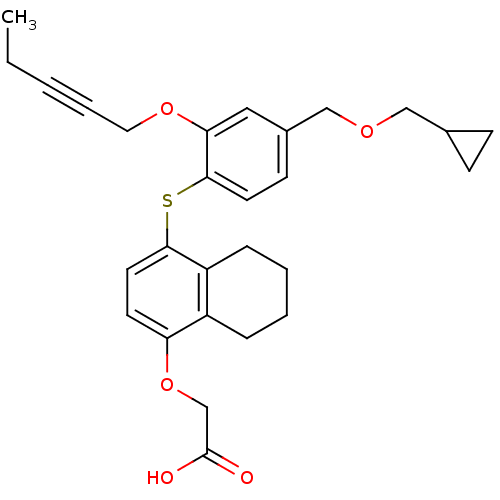
Inhibitor
Name:
BDBM29895
Synonyms:
alkynyl ether, 37
Type:
Small organic molecule
Emp. Form.:
C28H32O5S
Mol. Mass.:
480.616
SMILES:
CCC#CCOc1cc(COCC2CC2)ccc1Sc1ccc(OCC(O)=O)c2CCCCc12
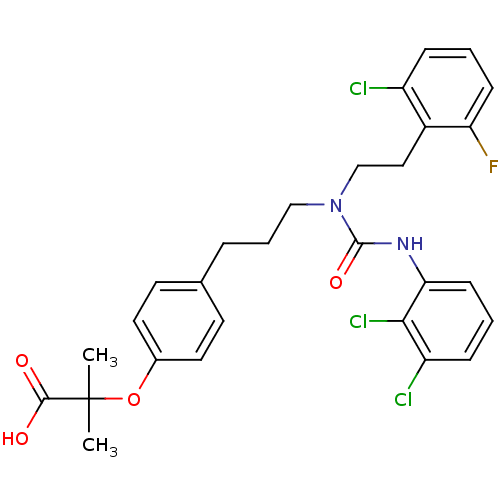
Substrate
Name:
BDBM28662
Synonyms:
2-[4-(3-{[2-(2-chloro-6-fluorophenyl)ethyl][(2,3-dichlorophenyl)carbamoyl]amino}propyl)phenoxy]-2-methylpropanoic acid | CHEMBL435278 | [3H]GW 2433
Type:
radiolabeled ligand
Emp. Form.:
C28H28Cl3FN2O4
Mol. Mass.:
581.89
SMILES:
CC(C)(Oc1ccc(CCCN(CCc2c(F)cccc2Cl)C(=O)Nc2cccc(Cl)c2Cl)cc1)C(O)=O