Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Insulin-degrading enzyme
Ligand
BDBM75814
Substrate
n/a
Meas. Tech.
Dose Response: Fluorescence polarization-based cell-based high throughput dose response assay for inhibitors of insulin-degrading enzyme (IDE)
IC50
5433±n/a nM
Citation
 PubChem, PC Dose Response: Fluorescence polarization-based cell-based high throughput dose response assay for inhibitors of insulin-degrading enzyme (IDE) PubChem Bioassay (2010)[AID]
PubChem, PC Dose Response: Fluorescence polarization-based cell-based high throughput dose response assay for inhibitors of insulin-degrading enzyme (IDE) PubChem Bioassay (2010)[AID] More Info.:
Target
Name:
Insulin-degrading enzyme
Synonyms:
Abeta-degrading protease | Human Insulin Degrading Enzyme (hIDE) | IDE | IDE_HUMAN | Insulin degrading enzyme (hIDE) | Insulin protease | Insulin-degrading enzyme | Insulinase | Insulysin | insulin-degrading enzyme isoform 1 precursor
Type:
Enzyme Catalytic Domain
Mol. Mass.:
117968.59
Organism:
Homo sapiens (Human)
Description:
P14735
Residue:
1019
Sequence:
MRYRLAWLLHPALPSTFRSVLGARLPPPERLCGFQKKTYSKMNNPAIKRIGNHITKSPEDKREYRGLELANGIKVLLISDPTTDKSSAALDVHIGSLSDPPNIAGLSHFCEHMLFLGTKKYPKENEYSQFLSEHAGSSNAFTSGEHTNYYFDVSHEHLEGALDRFAQFFLCPLFDESCKDREVNAVDSEHEKNVMNDAWRLFQLEKATGNPKHPFSKFGTGNKYTLETRPNQEGIDVRQELLKFHSAYYSSNLMAVCVLGRESLDDLTNLVVKLFSEVENKNVPLPEFPEHPFQEEHLKQLYKIVPIKDIRNLYVTFPIPDLQKYYKSNPGHYLGHLIGHEGPGSLLSELKSKGWVNTLVGGQKEGARGFMFFIINVDLTEEGLLHVEDIILHMFQYIQKLRAEGPQEWVFQECKDLNAVAFRFKDKERPRGYTSKIAGILHYYPLEEVLTAEYLLEEFRPDLIEMVLDKLRPENVRVAIVSKSFEGKTDRTEEWYGTQYKQEAIPDEVIKKWQNADLNGKFKLPTKNEFIPTNFEILPLEKEATPYPALIKDTAMSKLWFKQDDKFFLPKACLNFEFFSPFAYVDPLHCNMAYLYLELLKDSLNEYAYAAELAGLSYDLQNTIYGMYLSVKGYNDKQPILLKKIIEKMATFEIDEKRFEIIKEAYMRSLNNFRAEQPHQHAMYYLRLLMTEVAWTKDELKEALDDVTLPRLKAFIPQLLSRLHIEALLHGNITKQAALGIMQMVEDTLIEHAHTKPLLPSQLVRYREVQLPDRGWFVYQQRNEVHNNCGIEIYYQTDMQSTSENMFLELFCQIISEPCFNTLRTKEQLGYIVFSGPRRANGIQGLRFIIQSEKPPHYLESRVEAFLITMEKSIEDMTEEAFQKHIQALAIRRLDKPKKLSAECAKYWGEIISQQYNFDRDNTEVAYLKTLTKEDIIKFYKEMLAVDAPRRHKVSVHVLAREMDSCPVVGEFPCQNDINLSQAPALPQPEVIQNMTEFKRGLPLFPLVKPHINFMAAKL
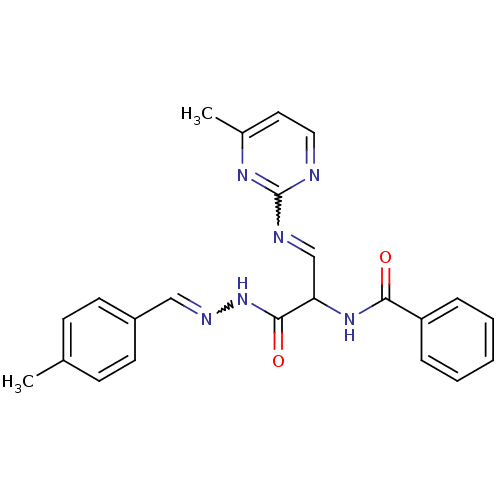
Inhibitor
Name:
BDBM75814
Synonyms:
MLS000539595 | N-[(E)-1-[[(4-methylbenzylidene)amino]carbamoyl]-2-[(4-methylpyrimidin-2-yl)amino]vinyl]benzamide | N-[(E)-3-[2-[(4-methylphenyl)methylidene]hydrazinyl]-1-[(4-methyl-2-pyrimidinyl)amino]-3-oxoprop-1-en-2-yl]benzamide | N-[(E)-3-[2-[(4-methylphenyl)methylidene]hydrazinyl]-1-[(4-methylpyrimidin-2-yl)amino]-3-oxidanylidene-prop-1-en-2-yl]benzamide | N-[(E)-3-[2-[(4-methylphenyl)methylidene]hydrazinyl]-1-[(4-methylpyrimidin-2-yl)amino]-3-oxoprop-1-en-2-yl]benzamide | N-{1-({2-[(4-methylphenyl)methylene]hydrazino}carbonyl)-2-[(4-methyl-2-pyrimidinyl)amino]vinyl}benzenecarboxamide | SMR000125253 | cid_2768717
Type:
Small organic molecule
Emp. Form.:
C23H22N6O2
Mol. Mass.:
414.4598
SMILES:
Cc1ccc(C=NNC(=O)C(NC(=O)c2ccccc2)C=Nc2nccc(C)n2)cc1 |w:21.22,6.6|