Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Fatty-acid amide hydrolase 1 [30-579]
Ligand
BDBM26739
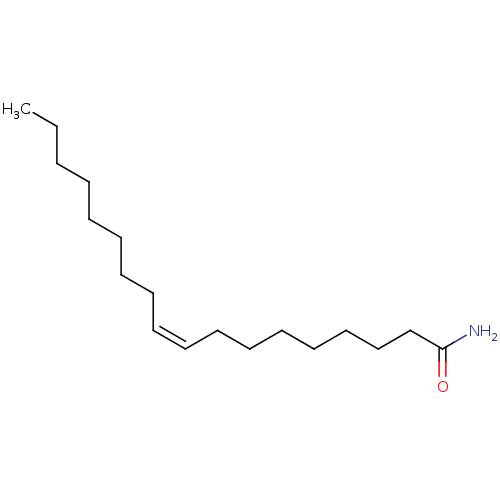
Substrate
BDBM23028
Meas. Tech.
Enzyme-coupled Assay
Ki
2.0e+3± 3e+2 nM
Citation
 Ahn, K; Johnson, DS; Mileni, M; Beidler, D; Long, JZ; McKinney, MK; Weerapana, E; Sadagopan, N; Liimatta, M; Smith, SE; Lazerwith, S; Stiff, C; Kamtekar, S; Bhattacharya, K; Zhang, Y; Swaney, S; Van Becelaere, K; Stevens, RC; Cravatt, BF Discovery and characterization of a highly selective FAAH inhibitor that reduces inflammatory pain. Chem Biol 16:411-20 (2009) [PubMed] Article
Ahn, K; Johnson, DS; Mileni, M; Beidler, D; Long, JZ; McKinney, MK; Weerapana, E; Sadagopan, N; Liimatta, M; Smith, SE; Lazerwith, S; Stiff, C; Kamtekar, S; Bhattacharya, K; Zhang, Y; Swaney, S; Van Becelaere, K; Stevens, RC; Cravatt, BF Discovery and characterization of a highly selective FAAH inhibitor that reduces inflammatory pain. Chem Biol 16:411-20 (2009) [PubMed] Article More Info.:
Target
Name:
Fatty-acid amide hydrolase 1 [30-579]
Synonyms:
Anandamide amidohydrolase 1 | FAAH1_RAT | Faah | Faah1 | Fatty Acid Amide Hydrolase | Fatty Acid Amide Hydrolic, FAAH | Fatty-acid amide hydrolase (FAAH) | Fatty-acid amide hydrolase 1 | Fatty-acid amide hydrolase 1 (FAAH) | Fatty-acid amide hydrolase 1 (aa 30-579) | Oleamide hydrolase 1
Type:
Single-pass membrane protein; homodimer
Mol. Mass.:
60474.00
Organism:
Rattus norvegicus (rat)
Description:
P97612 (aa 30-579)
Residue:
550
Sequence:
RWTGRQKARGAATRARQKQRASLETMDKAVQRFRLQNPDLDSEALLTLPLLQLVQKLQSGELSPEAVFFTYLGKAWEVNKGTNCVTSYLTDCETQLSQAPRQGLLYGVPVSLKECFSYKGHDSTLGLSLNEGMPSESDCVVVQVLKLQGAVPFVHTNVPQSMLSFDCSNPLFGQTMNPWKSSKSPGGSSGGEGALIGSGGSPLGLGTDIGGSIRFPSAFCGICGLKPTGNRLSKSGLKGCVYGQTAVQLSLGPMARDVESLALCLKALLCEHLFTLDPTVPPLPFREEVYRSSRPLRVGYYETDNYTMPSPAMRRALIETKQRLEAAGHTLIPFLPNNIPYALEVLSAGGLFSDGGRSFLQNFKGDFVDPCLGDLILILRLPSWFKRLLSLLLKPLFPRLAAFLNSMRPRSAEKLWKLQHEIEMYRQSVIAQWKAMNLDVLLTPMLGPALDLNTPGRATGAISYTVLYNCLDFPAGVVPVTTVTAEDDAQMELYKGYFGDIWDIILKKAMKNSVGLPVAVQCVALPWQEELCLRFMREVEQLMTPQKQPS
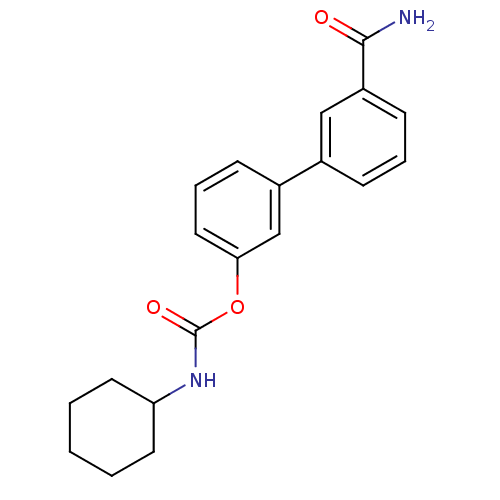
Inhibitor
Name:
BDBM26739
Synonyms:
3-(3-carbamoylphenyl)phenyl N-cyclohexylcarbamate | CHEMBL184238 | URB 597 | URB-597 | URB597 | US9187413, 1a (URB597)
Type:
Small organic molecule
Emp. Form.:
C20H22N2O3
Mol. Mass.:
338.4003
SMILES:
NC(=O)c1cccc(c1)-c1cccc(OC(=O)NC2CCCCC2)c1