Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Alpha-1A adrenergic receptor
Ligand
BDBM50001888
Substrate
n/a
Ki
14±n/a nM
Comments
PDSP_513
Citation
 Hall, H; Sällemark, M; Jerning, E Effects of remoxipride and some related new substituted salicylamides on rat brain receptors. Acta Pharmacol Toxicol (Copenh) 58:61-70 (1986) [PubMed] Article
Hall, H; Sällemark, M; Jerning, E Effects of remoxipride and some related new substituted salicylamides on rat brain receptors. Acta Pharmacol Toxicol (Copenh) 58:61-70 (1986) [PubMed] Article More Info.:
Target
Name:
Alpha-1A adrenergic receptor
Synonyms:
ADA1A_RAT | Adra1a | Adra1c | Alpha-1 adrenoreceptor | Alpha-1A adrenoceptor | Alpha-1A adrenoreceptor | Alpha-1C adrenergic receptor | adrenergic Alpha1A
Type:
Protein
Mol. Mass.:
51620.15
Organism:
Rattus norvegicus (Rat)
Description:
P43140
Residue:
466
Sequence:
MVLLSENASEGSNCTHPPAPVNISKAILLGVILGGLIIFGVLGNILVILSVACHRHLHSVTHYYIVNLAVADLLLTSTVLPFSAIFEILGYWAFGRVFCNIWAAVDVLCCTASIMGLCIISIDRYIGVSYPLRYPTIVTQRRGVRALLCVWVLSLVISIGPLFGWRQPAPEDETICQINEEPGYVLFSALGSFYVPLAIILVMYCRVYVVAKRESRGLKSGLKTDKSDSEQVTLRIHRKNVPAEGGGVSSAKNKTHFSVRLLKFSREKKAAKTLGIVVGCFVLCWLPFFLVMPIGSFFPDFKPSETVFKIVFWLGYLNSCINPIIYPCSSQEFKKAFQNVLRIQCLRRRQSSKHALGYTLHPPSQALEGQHRDMVRIPVGSGETFYKISKTDGVCEWKFFSSMPQGSARITVPKDQSACTTARVRSKSFLQVCCCVGSSAPRPEENHQVPTIKIHTISLGENGEEV
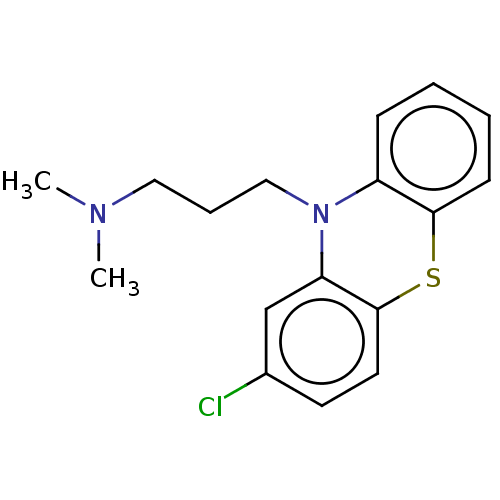
Inhibitor
Name:
BDBM50001888
Synonyms:
(chloropromazine) [3-(2-Chloro-phenothiazin-10-yl)-propyl]-dimethyl-amine | (chlorpromazine)[3-(2-Chloro-phenothiazin-10-yl)-propyl]-dimethyl-amine | 1-(2-Allyl-phenoxy)-3-isopropylamino-propan-2-ol | 1N,1N-dimethyl-3-(2-chloro-10H-10-phenothiazinyl)-1-propanamine | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine | CHEMBL71 | CHLORPROMAZINE | CHLORPROMAZINE HIBENZATE | CHLORPROMAZINE HYDROCHLORIDE | CHLORPROMAZINE PHENOLPHTHALINATE | CHLORPROMAZINE TANNATE | Chlorpromazine;[3-(2-Chloro-phenothiazin-10-yl)-propyl]-dimethyl-amine | PROMAPAR | SONAZINE | THORAZINE | [3-(2-Chloro-phenothiazin-10-yl)-propyl]-dimethyl-amine (chlor-promazine) | [3-(2-Chloro-phenothiazin-10-yl)-propyl]-dimethyl-amine( Chlorpromazine) | [3-(2-Chloro-phenothiazin-10-yl)-propyl]-dimethyl-amine(clorpromazine) | chloropromazine | med.21724, Compound 15
Type:
Small organic molecule
Emp. Form.:
C17H19ClN2S
Mol. Mass.:
318.864
SMILES:
CN(C)CCCN1c2ccccc2Sc2ccc(Cl)cc12