Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
MAP kinase-interacting serine/threonine-protein kinase 1
Ligand
BDBM50263264
Substrate
n/a
Meas. Tech.
ChEBML_1697749
IC50
1.5±n/a nM
Citation
 Reich, SH; Sprengeler, PA; Chiang, GG; Appleman, JR; Chen, J; Clarine, J; Eam, B; Ernst, JT; Han, Q; Goel, VK; Han, EZR; Huang, V; Hung, INJ; Jemison, A; Jessen, KA; Molter, J; Murphy, D; Neal, M; Parker, GS; Shaghafi, M; Sperry, S; Staunton, J; Stumpf, CR; Thompson, PA; Tran, C; Webber, SE; Wegerski, CJ; Zheng, H; Webster, KR Structure-based Design of Pyridone-Aminal eFT508 Targeting Dysregulated Translation by Selective Mitogen-activated Protein Kinase Interacting Kinases 1 and 2 (MNK1/2) Inhibition. J Med Chem 61:3516-3540 (2018) [PubMed] Article
Reich, SH; Sprengeler, PA; Chiang, GG; Appleman, JR; Chen, J; Clarine, J; Eam, B; Ernst, JT; Han, Q; Goel, VK; Han, EZR; Huang, V; Hung, INJ; Jemison, A; Jessen, KA; Molter, J; Murphy, D; Neal, M; Parker, GS; Shaghafi, M; Sperry, S; Staunton, J; Stumpf, CR; Thompson, PA; Tran, C; Webber, SE; Wegerski, CJ; Zheng, H; Webster, KR Structure-based Design of Pyridone-Aminal eFT508 Targeting Dysregulated Translation by Selective Mitogen-activated Protein Kinase Interacting Kinases 1 and 2 (MNK1/2) Inhibition. J Med Chem 61:3516-3540 (2018) [PubMed] Article More Info.:
Target
Name:
MAP kinase-interacting serine/threonine-protein kinase 1
Synonyms:
MAP Kinase-Interacting Protein Kinase (MNK1) | MAP kinase signal-integrating kinase 1 | MAP kinase-interacting serine/threonine-protein kinase 1 (MnK1) | MAP kinase-interacting serine/threonine-protein kinase MNK1 | MAP-kinase interacting kinase 1 (MNK1) | MAPK signal-integrating kinase 1 | MKNK1 | MKNK1_HUMAN | MNK1
Type:
Serine/threonine-protein kinase
Mol. Mass.:
51342.85
Organism:
Homo sapiens (Human)
Description:
Q9BUB5
Residue:
465
Sequence:
MVSSQKLEKPIEMGSSEPLPIADGDRRRKKKRRGRATDSLPGKFEDMYKLTSELLGEGAYAKVQGAVSLQNGKEYAVKIIEKQAGHSRSRVFREVETLYQCQGNKNILELIEFFEDDTRFYLVFEKLQGGSILAHIQKQKHFNEREASRVVRDVAAALDFLHTKDKVSLCHLGWSAMAPSGLTAAPTSLGSSDPPTSASQVAGTTGIAHRDLKPENILCESPEKVSPVKICDFDLGSGMKLNNSCTPITTPELTTPCGSAEYMAPEVVEVFTDQATFYDKRCDLWSLGVVLYIMLSGYPPFVGHCGADCGWDRGEVCRVCQNKLFESIQEGKYEFPDKDWAHISSEAKDLISKLLVRDAKQRLSAAQVLQHPWVQGQAPEKGLPTPQVLQRNSSTMDLTLFAAEAIALNRQLSQHEENELAEEPEALADGLCSMKLSPPCKSRLARRRALAQAGRGEDRSPPTAL
Inhibitor
Name:
BDBM50263264
Synonyms:
CHEMBL4094080
Type:
Small organic molecule
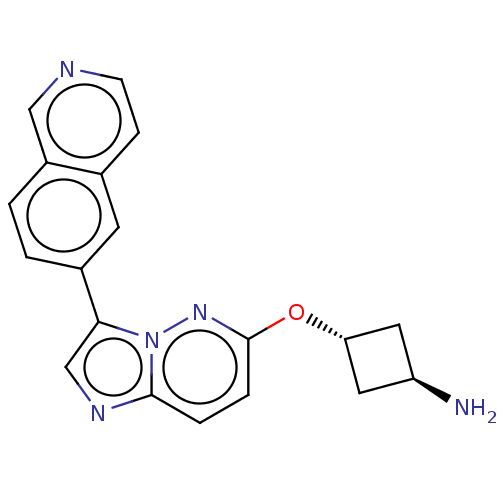
Emp. Form.:
C19H17N5O
Mol. Mass.:
331.3712
SMILES:
N[C@H]1C[C@@H](C1)Oc1ccc2ncc(-c3ccc4cnccc4c3)n2n1 |r,wU:3.5,wD:1.0,(36.72,-33.01,;35.21,-33.32,;33.93,-32.47,;33.08,-33.76,;34.37,-34.61,;31.57,-34.07,;30.55,-32.92,;29.04,-33.23,;28.03,-32.08,;28.51,-30.62,;27.74,-29.3,;28.77,-28.15,;30.18,-28.78,;31.51,-28.01,;31.51,-26.46,;32.84,-25.69,;34.17,-26.46,;35.5,-25.68,;36.83,-26.44,;36.84,-27.99,;35.51,-28.77,;34.17,-28,;32.84,-28.78,;30.01,-30.31,;31.04,-31.45,)|