Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Matrix metalloproteinase-9
Ligand
BDBM50128660
Substrate
n/a
Meas. Tech.
ChEMBL_105516 (CHEMBL715225)
IC50
13±n/a nM
Citation
 Aranapakam, V; Davis, JM; Grosu, GT; Baker, J; Ellingboe, J; Zask, A; Levin, JI; Sandanayaka, VP; Du, M; Skotnicki, JS; DiJoseph, JF; Sung, A; Sharr, MA; Killar, LM; Walter, T; Jin, G; Cowling, R; Tillett, J; Zhao, W; McDevitt, J; Xu, ZB Synthesis and structure-activity relationship of N-substituted 4-arylsulfonylpiperidine-4-hydroxamic acids as novel, orally active matrix metalloproteinase inhibitors for the treatment of osteoarthritis. J Med Chem 46:2376-96 (2003) [PubMed] Article
Aranapakam, V; Davis, JM; Grosu, GT; Baker, J; Ellingboe, J; Zask, A; Levin, JI; Sandanayaka, VP; Du, M; Skotnicki, JS; DiJoseph, JF; Sung, A; Sharr, MA; Killar, LM; Walter, T; Jin, G; Cowling, R; Tillett, J; Zhao, W; McDevitt, J; Xu, ZB Synthesis and structure-activity relationship of N-substituted 4-arylsulfonylpiperidine-4-hydroxamic acids as novel, orally active matrix metalloproteinase inhibitors for the treatment of osteoarthritis. J Med Chem 46:2376-96 (2003) [PubMed] Article More Info.:
Target
Name:
Matrix metalloproteinase-9
Synonyms:
67 kDa matrix metalloproteinase-9 | 82 kDa matrix metalloproteinase-9 | 92 kDa gelatinase | 92 kDa type IV collagenase | CLG4B | GELB | Gelatinase B | MMP-9 | MMP9 | MMP9_HUMAN | Matrix metalloproteinase 9 (MMP-9) | Matrix metalloproteinase-9 (MMP-9) | Matrix metalloproteinase-9 (MMP9)
Type:
Enzyme
Mol. Mass.:
78452.28
Organism:
Homo sapiens (Human)
Description:
P14780
Residue:
707
Sequence:
MSLWQPLVLVLLVLGCCFAAPRQRQSTLVLFPGDLRTNLTDRQLAEEYLYRYGYTRVAEMRGESKSLGPALLLLQKQLSLPETGELDSATLKAMRTPRCGVPDLGRFQTFEGDLKWHHHNITYWIQNYSEDLPRAVIDDAFARAFALWSAVTPLTFTRVYSRDADIVIQFGVAEHGDGYPFDGKDGLLAHAFPPGPGIQGDAHFDDDELWSLGKGVVVPTRFGNADGAACHFPFIFEGRSYSACTTDGRSDGLPWCSTTANYDTDDRFGFCPSERLYTQDGNADGKPCQFPFIFQGQSYSACTTDGRSDGYRWCATTANYDRDKLFGFCPTRADSTVMGGNSAGELCVFPFTFLGKEYSTCTSEGRGDGRLWCATTSNFDSDKKWGFCPDQGYSLFLVAAHEFGHALGLDHSSVPEALMYPMYRFTEGPPLHKDDVNGIRHLYGPRPEPEPRPPTTTTPQPTAPPTVCPTGPPTVHPSERPTAGPTGPPSAGPTGPPTAGPSTATTVPLSPVDDACNVNIFDAIAEIGNQLYLFKDGKYWRFSEGRGSRPQGPFLIADKWPALPRKLDSVFEERLSKKLFFFSGRQVWVYTGASVLGPRRLDKLGLGADVAQVTGALRSGRGKMLLFSGRRLWRFDVKAQMVDPRSASEVDRMFPGVPLDTHDVFQYREKAYFCQDRFYWRVSSRSELNQVDQVGYVTYDILQCPED
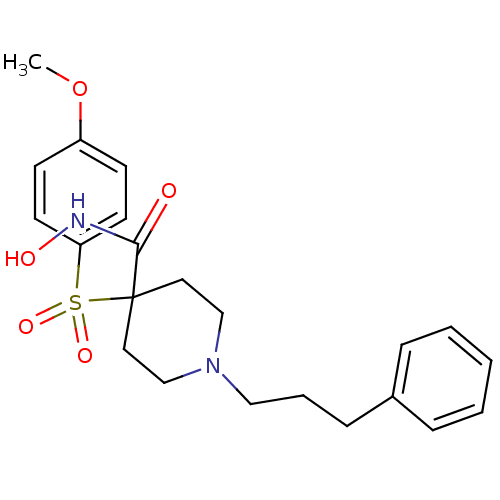
Inhibitor
Name:
BDBM50128660
Synonyms:
4-(4-Methoxy-benzenesulfonyl)-1-(3-phenyl-propyl)-piperidine-4-carboxylic acid hydroxyamide | CHEMBL312770 | N-hydroxy-4-(4-methoxyphenylsulfonyl)-1-(3-phenylpropyl)piperidine-4-carboxamide
Type:
Small organic molecule
Emp. Form.:
C22H28N2O5S
Mol. Mass.:
432.533
SMILES:
COc1ccc(cc1)S(=O)(=O)C1(CCN(CCCc2ccccc2)CC1)C(=O)NO