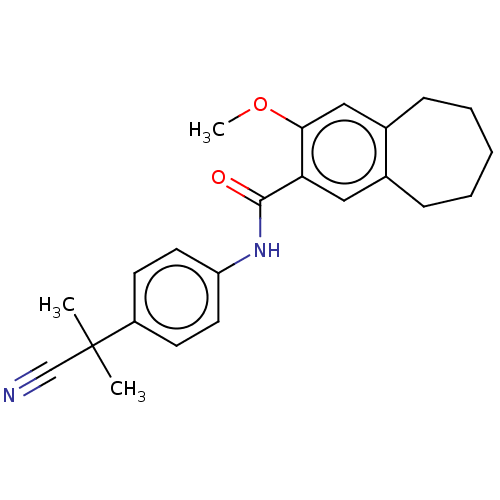
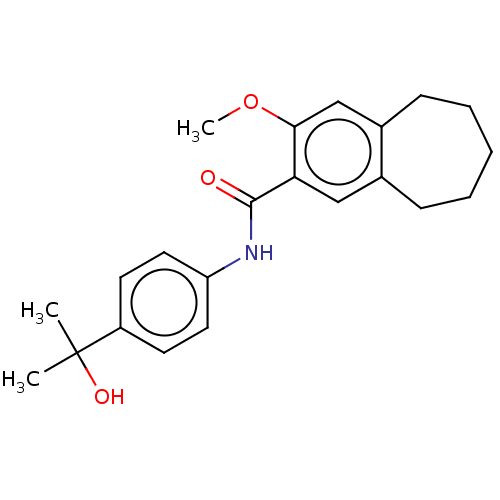
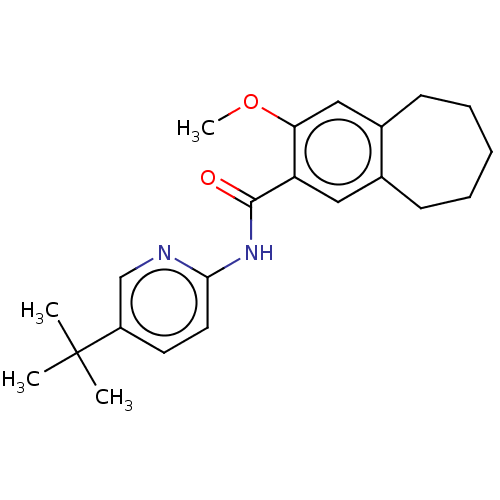
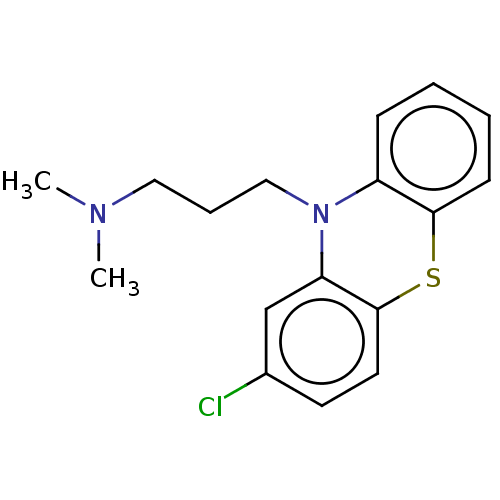
TargetSodium-dependent serotonin transporter(Rattus norvegicus (rat))
Central Washington University
Curated by ChEMBL
Central Washington University
Curated by ChEMBL
Affinity DataKi: 0.0810nMAssay Description:In vitro radioligand [3H]-paroxetine from rat cortical Serotonin transporterMore data for this Ligand-Target Pair
TargetSodium-dependent serotonin transporter(Rattus norvegicus (rat))
Central Washington University
Curated by ChEMBL
Central Washington University
Curated by ChEMBL
Affinity DataKi: 0.163nMAssay Description:In vitro radioligand [3H]-paroxetine from rat cortical Serotonin transporterMore data for this Ligand-Target Pair
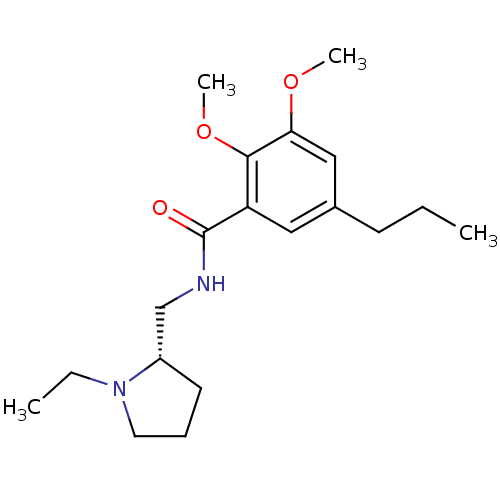
TargetSodium-dependent serotonin transporter(Rattus norvegicus (rat))
Central Washington University
Curated by ChEMBL
Central Washington University
Curated by ChEMBL
Affinity DataKi: 0.163nMAssay Description:In vitro radioligand [3H]-paroxetine from rat cortical Serotonin transporterMore data for this Ligand-Target Pair
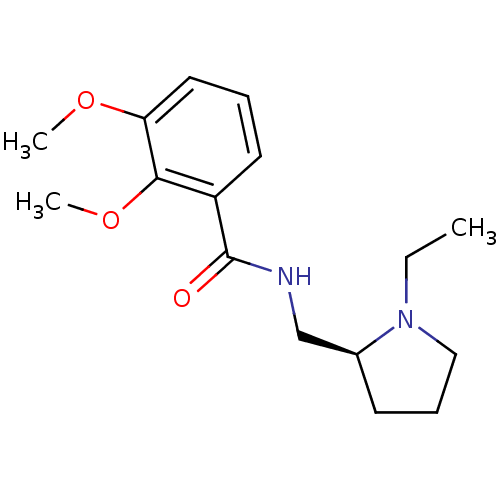
TargetSodium-dependent serotonin transporter(Rattus norvegicus (rat))
Central Washington University
Curated by ChEMBL
Central Washington University
Curated by ChEMBL
Affinity DataKi: 0.190nMAssay Description:In vitro radioligand [3H]-paroxetine from rat cortical Serotonin transporterMore data for this Ligand-Target Pair
Affinity DataKi: 0.190nMAssay Description:Compound was evaluated for binding affinity towards dopamine receptor D2 in striatal membranes, using [3H]- spiperone as radioligand in the presence ...More data for this Ligand-Target Pair
Affinity DataKi: 0.200nMAssay Description:Compound was evaluated for binding affinity towards dopamine receptor D2 in striatal membranes, using [3H]- spiperone as radioligand in the presence ...More data for this Ligand-Target Pair
Affinity DataKi: 0.25nMAssay Description:Compound was evaluated for binding affinity towards dopamine receptor D2 in striatal membranes, using [3H]- spiperone as radioligand in the presence ...More data for this Ligand-Target Pair
Affinity DataKi: 0.270nMAssay Description:Compound was evaluated for binding affinity towards Dopamine receptor D2 in striatal membranes, using [3H]- spiperone as radioligand in the absence o...More data for this Ligand-Target Pair
Affinity DataKi: 0.660nMAssay Description:Displacement of [3H]- spiperone from dopamine receptor D2 of striatal membranes without sodium chlorideMore data for this Ligand-Target Pair
Affinity DataKi: 0.760nMAssay Description:Displacement of [3H]- spiperone from dopamine receptor D2 of striatal membranes without sodium chlorideMore data for this Ligand-Target Pair
Affinity DataKi: 0.770nMAssay Description:Compound was evaluated for binding affinity towards dopamine receptor D2 in striatal membranes, using [3H]- spiperone as radioligand in the presence ...More data for this Ligand-Target Pair
Affinity DataKi: 0.770nMAssay Description:Displacement of [3H]- spiperone from dopamine receptor D2 of striatal membranes without sodium chlorideMore data for this Ligand-Target Pair
Affinity DataKi: 0.800nMAssay Description:Displacement of [3H]- spiperone from dopamine receptor D2 of striatal membranes without sodium chlorideMore data for this Ligand-Target Pair
Affinity DataKi: 0.990nMAssay Description:Displacement of [3H]- spiperone from dopamine receptor D2 of striatal membranes without sodium chlorideMore data for this Ligand-Target Pair
Affinity DataKi: 1.20nMAssay Description:Compound was evaluated for binding affinity towards dopamine receptor D2 in striatal membranes, using [3H]- spiperone as radioligand in the presence ...More data for this Ligand-Target Pair
Affinity DataKi: 1.30nMAssay Description:Displacement of [3H]- spiperone from dopamine receptor D2 of striatal membranes without sodium chlorideMore data for this Ligand-Target Pair
TargetAlpha-2A adrenergic receptor [16-465]/Alpha-2B adrenergic receptor/Alpha-2C adrenergic receptor(RAT)
University of California
Curated by ChEMBL
University of California
Curated by ChEMBL
Affinity DataKi: 1.40nMAssay Description:Compound was evaluated for binding affinity towards dopamine receptor D2 in striatal membranes, using [3H]- spiperone as radioligand in the presence ...More data for this Ligand-Target Pair
Affinity DataKi: 1.40nMAssay Description:Compound was evaluated for binding affinity towards dopamine receptor D2 in striatal membranes, using [3H]- spiperone as radioligand in the presence ...More data for this Ligand-Target Pair
Affinity DataKi: 1.80nMAssay Description:Compound was evaluated for binding affinity towards Dopamine receptor D2 in striatal membranes, using [3H]- spiperone as radioligand in the absence o...More data for this Ligand-Target Pair
Affinity DataKi: 2.20nMAssay Description:Compound was evaluated for binding affinity towards dopamine receptor D2 in striatal membranes, using [3H]- spiperone as radioligand in the presence ...More data for this Ligand-Target Pair
Affinity DataKi: 4.40nMAssay Description:Compound was evaluated for binding affinity towards dopamine receptor D2 in striatal membranes, using [3H]- spiperone as radioligand in the presence ...More data for this Ligand-Target Pair
TargetSodium-dependent serotonin transporter(Rattus norvegicus (rat))
Central Washington University
Curated by ChEMBL
Central Washington University
Curated by ChEMBL
Affinity DataKi: 4.60nMAssay Description:In vitro radioligand [3H]-paroxetine from rat cortical Serotonin transporterMore data for this Ligand-Target Pair
TargetAlpha-2A adrenergic receptor [16-465]/Alpha-2B adrenergic receptor/Alpha-2C adrenergic receptor(RAT)
University of California
Curated by ChEMBL
University of California
Curated by ChEMBL
Affinity DataKi: 5nMAssay Description:Compound was evaluated for binding affinity towards dopamine receptor D2 in striatal membranes, using [3H]- spiperone as radioligand in the presence ...More data for this Ligand-Target Pair
Affinity DataKi: 5.30nMAssay Description:Displacement of [3H]- spiperone from dopamine receptor D2 of striatal membranes without sodium chlorideMore data for this Ligand-Target Pair
Affinity DataKi: 7nMAssay Description:Displacement of [3H]- spiperone from dopamine receptor D2 of striatal membranes without sodium chlorideMore data for this Ligand-Target Pair
Affinity DataKi: 8.30nMAssay Description:Displacement of [3H]- spiperone from dopamine receptor D2 of striatal membranes without sodium chlorideMore data for this Ligand-Target Pair
TargetAlpha-2A adrenergic receptor [16-465]/Alpha-2B adrenergic receptor/Alpha-2C adrenergic receptor(RAT)
University of California
Curated by ChEMBL
University of California
Curated by ChEMBL
Affinity DataKi: 9.5nMAssay Description:binding affinity towards alpha-2 adrenergic receptor, using [3H]- atipamezole as radioligand from rat frontal cortex membranesMore data for this Ligand-Target Pair
TargetAlpha-2A adrenergic receptor [16-465]/Alpha-2B adrenergic receptor/Alpha-2C adrenergic receptor(RAT)
University of California
Curated by ChEMBL
University of California
Curated by ChEMBL
Affinity DataKi: 11nMAssay Description:Compound was evaluated for binding affinity towards dopamine receptor D2 in striatal membranes, using [3H]- spiperone as radioligand in the presence ...More data for this Ligand-Target Pair
Affinity DataKi: 33nMAssay Description:Displacement of [3H]- spiperone from dopamine receptor D2 of striatal membranes without sodium chlorideMore data for this Ligand-Target Pair
Affinity DataKi: 66nMAssay Description:Displacement of [3H]- spiperone from dopamine receptor D2 of striatal membranes without sodium chlorideMore data for this Ligand-Target Pair
Affinity DataKi: >1.00E+4nMAssay Description:Displacement of [3H]- spiperone from dopamine receptor D2 of striatal membranes without sodium chlorideMore data for this Ligand-Target Pair
Affinity DataKi: >1.00E+4nMAssay Description:Displacement of [3H]- spiperone from dopamine receptor D2 of striatal membranes without sodium chlorideMore data for this Ligand-Target Pair
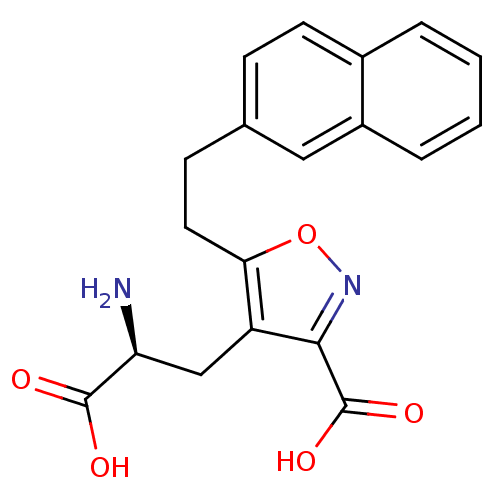
Affinity DataKi: 5.20E+4nMAssay Description:Competitive inhibition of [3H]L-glutamate uptake at amino acid transport system xc- in human SNB19 cells by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: 5.90E+4nMAssay Description:Competitive inhibition of [3H]L-glutamate uptake at amino acid transport system xc- in human SNB19 cells by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: 6.00E+4nMAssay Description:Competitive inhibition of [3H]L-glutamate uptake at amino acid transport system xc- in human SNB19 cells by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: 6.40E+4nMAssay Description:Competitive inhibition of [3H]L-glutamate uptake at amino acid transport system xc- in human SNB19 cells by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: 1.13E+5nMAssay Description:Competitive inhibition of [3H]L-glutamate uptake at amino acid transport system xc- in human SNB19 cells by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: 1.17E+5nMAssay Description:Competitive inhibition of [3H]L-glutamate uptake at amino acid transport system xc- in human SNB19 cells by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: 1.47E+5nMAssay Description:Competitive inhibition of [3H]L-glutamate uptake at amino acid transport system xc- in human SNB19 cells by liquid scintillation countingMore data for this Ligand-Target Pair
TargetDihydroorotate dehydrogenase (quinone), mitochondrial(Homo sapiens (Human))
Southern Research
Curated by ChEMBL
Southern Research
Curated by ChEMBL
Affinity DataIC50: 20nMAssay Description:Inhibition of human recombinant full length C-terminal MYC/DDk-tagged DHODH using decylubiquinone as substrate measured for 1 hr by DCIP absorbance a...More data for this Ligand-Target Pair
TargetDihydroorotate dehydrogenase (quinone), mitochondrial(Homo sapiens (Human))
Southern Research
Curated by ChEMBL
Southern Research
Curated by ChEMBL
Affinity DataIC50: 310nMAssay Description:Inhibition of human recombinant full length C-terminal MYC/DDk-tagged DHODH using decylubiquinone as substrate measured for 1 hr by DCIP absorbance a...More data for this Ligand-Target Pair
TargetDihydroorotate dehydrogenase (quinone), mitochondrial(Homo sapiens (Human))
Southern Research
Curated by ChEMBL
Southern Research
Curated by ChEMBL
Affinity DataIC50: 3.00E+3nMAssay Description:Inhibition of human recombinant full length C-terminal MYC/DDk-tagged DHODH using decylubiquinone as substrate measured for 1 hr by DCIP absorbance a...More data for this Ligand-Target Pair
TargetDihydroorotate dehydrogenase (quinone), mitochondrial(Homo sapiens (Human))
Southern Research
Curated by ChEMBL
Southern Research
Curated by ChEMBL
Affinity DataIC50: 3.50E+3nMAssay Description:Inhibition of human recombinant full length C-terminal MYC/DDk-tagged DHODH using decylubiquinone as substrate measured for 1 hr by DCIP absorbance a...More data for this Ligand-Target Pair
TargetDihydroorotate dehydrogenase (quinone), mitochondrial(Homo sapiens (Human))
Southern Research
Curated by ChEMBL
Southern Research
Curated by ChEMBL
Affinity DataIC50: 5.00E+3nMAssay Description:Inhibition of human recombinant full length C-terminal MYC/DDk-tagged DHODH using decylubiquinone as substrate measured for 1 hr by DCIP absorbance a...More data for this Ligand-Target Pair
TargetDihydroorotate dehydrogenase (quinone), mitochondrial(Homo sapiens (Human))
Southern Research
Curated by ChEMBL
Southern Research
Curated by ChEMBL
Affinity DataIC50: 2.41E+4nMAssay Description:Inhibition of human recombinant full length C-terminal MYC/DDk-tagged DHODH using decylubiquinone as substrate measured for 1 hr by DCIP absorbance a...More data for this Ligand-Target Pair
TargetDihydroorotate dehydrogenase (quinone), mitochondrial(Homo sapiens (Human))
Southern Research
Curated by ChEMBL
Southern Research
Curated by ChEMBL
Affinity DataIC50: >3.00E+4nMAssay Description:Inhibition of human recombinant full length C-terminal MYC/DDk-tagged DHODH using decylubiquinone as substrate measured for 1 hr by DCIP absorbance a...More data for this Ligand-Target Pair
TargetDihydroorotate dehydrogenase (quinone), mitochondrial(Homo sapiens (Human))
Southern Research
Curated by ChEMBL
Southern Research
Curated by ChEMBL
Affinity DataIC50: >3.00E+4nMAssay Description:Inhibition of human recombinant full length C-terminal MYC/DDk-tagged DHODH using decylubiquinone as substrate measured for 1 hr by DCIP absorbance a...More data for this Ligand-Target Pair
TargetDihydroorotate dehydrogenase (quinone), mitochondrial(Homo sapiens (Human))
Southern Research
Curated by ChEMBL
Southern Research
Curated by ChEMBL
Affinity DataIC50: >3.00E+4nMAssay Description:Inhibition of human recombinant full length C-terminal MYC/DDk-tagged DHODH using decylubiquinone as substrate measured for 1 hr by DCIP absorbance a...More data for this Ligand-Target Pair
TargetDihydroorotate dehydrogenase (quinone), mitochondrial(Homo sapiens (Human))
Southern Research
Curated by ChEMBL
Southern Research
Curated by ChEMBL
Affinity DataIC50: >3.00E+4nMAssay Description:Inhibition of human recombinant full length C-terminal MYC/DDk-tagged DHODH using decylubiquinone as substrate measured for 1 hr by DCIP absorbance a...More data for this Ligand-Target Pair
TargetDihydroorotate dehydrogenase (quinone), mitochondrial(Homo sapiens (Human))
Southern Research
Curated by ChEMBL
Southern Research
Curated by ChEMBL
Affinity DataIC50: >3.00E+4nMAssay Description:Inhibition of human recombinant full length C-terminal MYC/DDk-tagged DHODH using decylubiquinone as substrate measured for 1 hr by DCIP absorbance a...More data for this Ligand-Target Pair



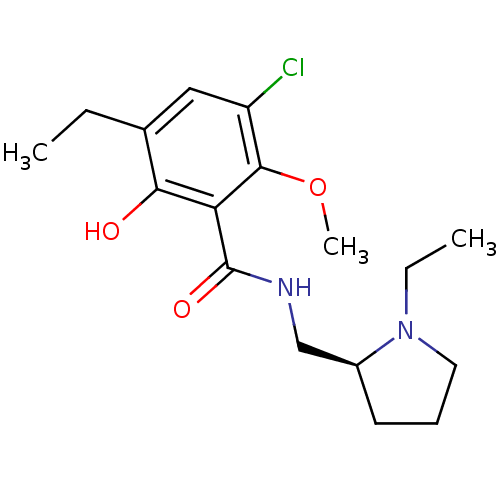


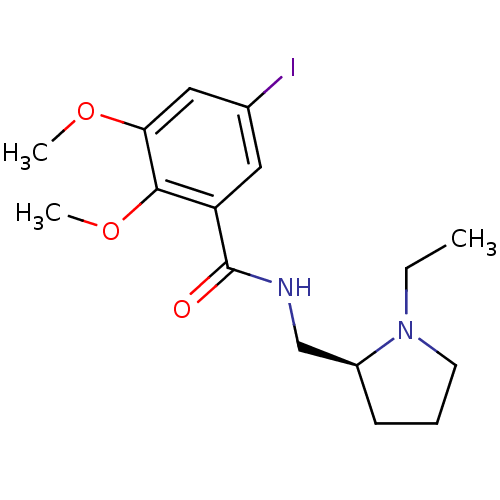

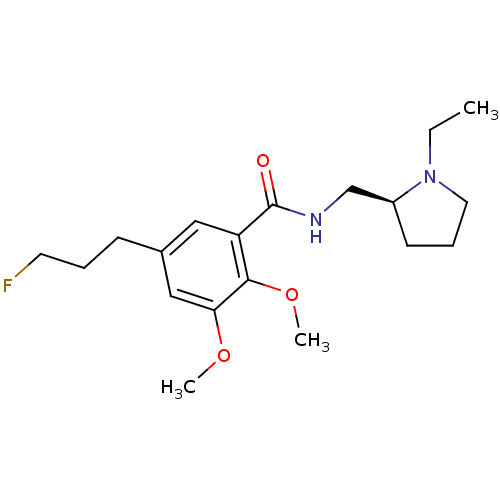

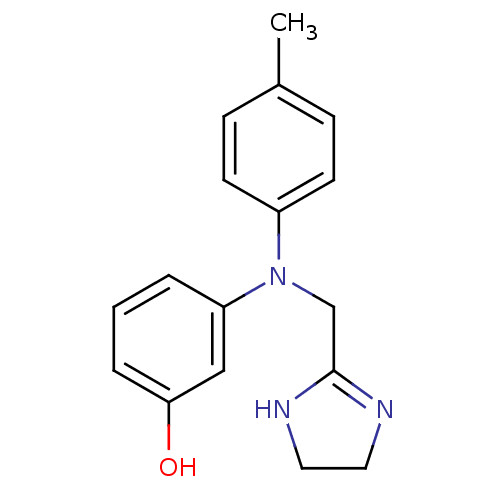










 3D Structure (crystal)
3D Structure (crystal)