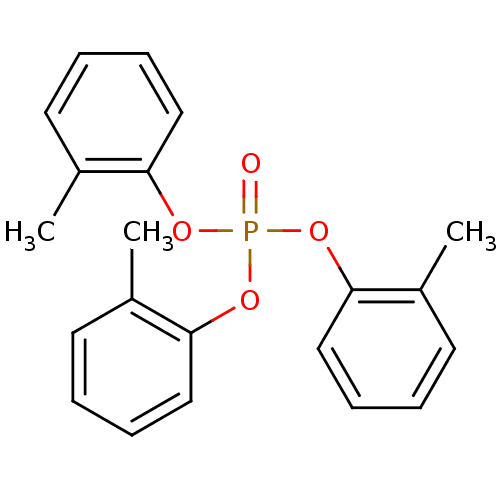
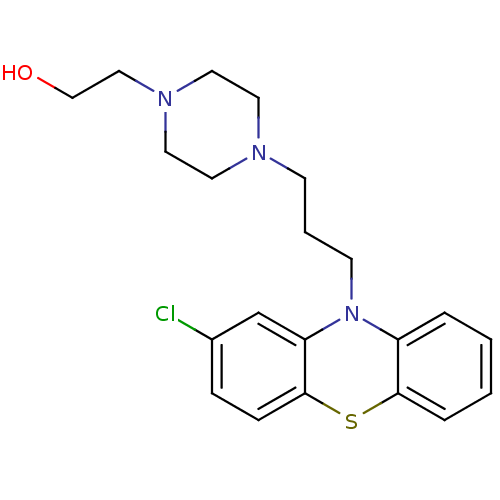
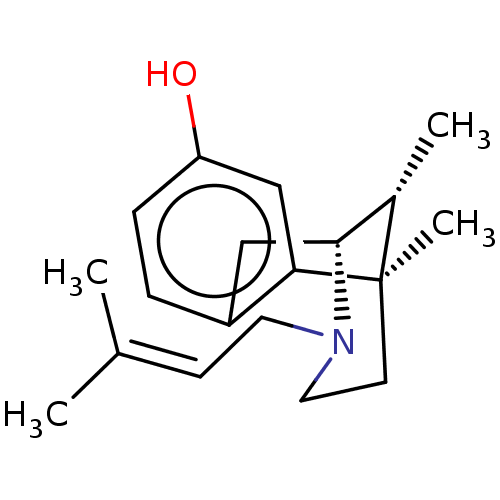
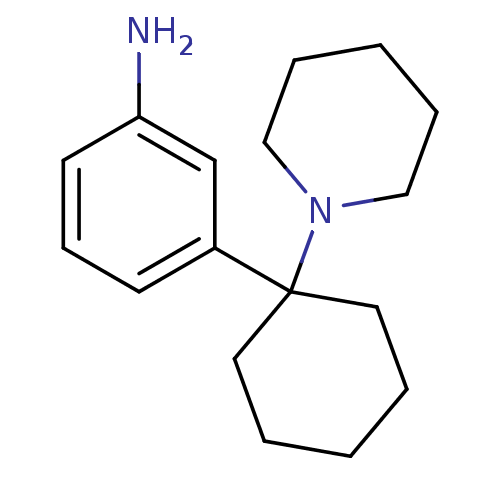
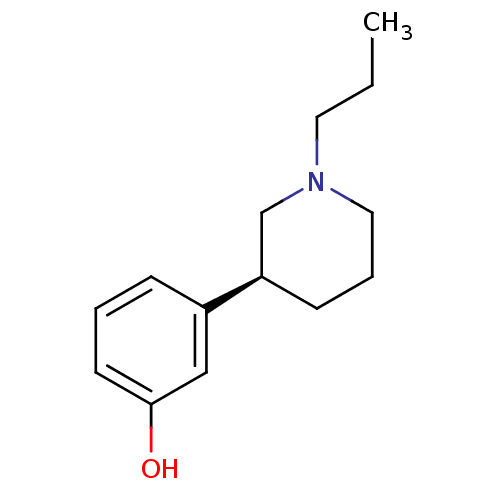
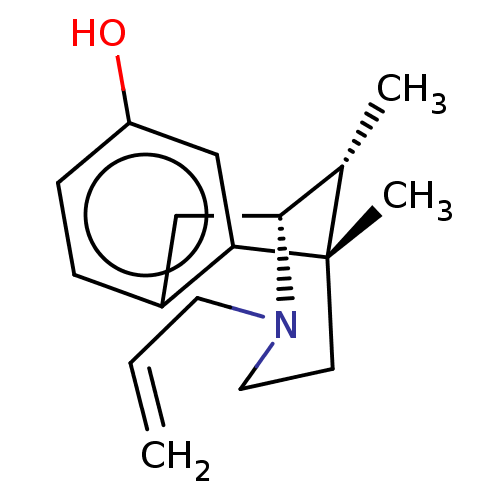
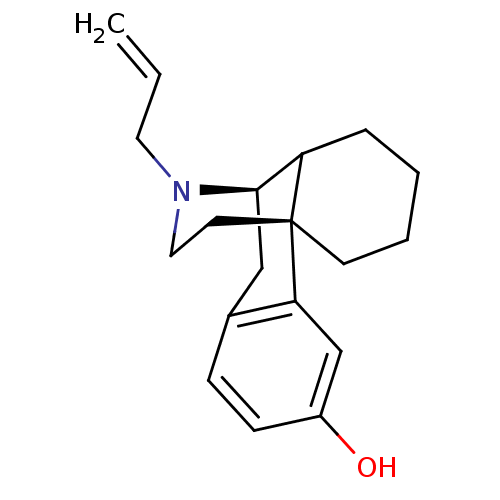
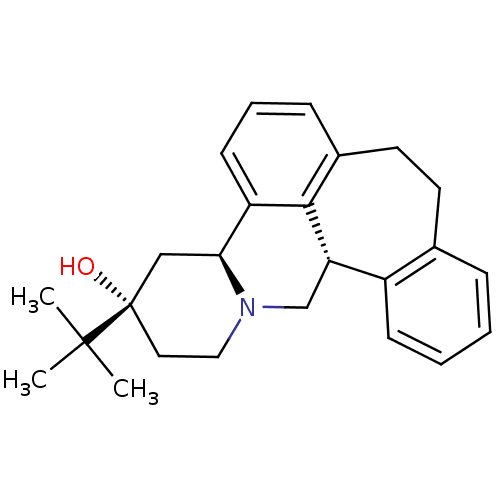
TargetGlutamate receptor ionotropic, NMDA 1(RAT)
The Johns Hopkins University
Curated by PDSP Ki Database
The Johns Hopkins University
Curated by PDSP Ki Database
TargetGlutamate receptor ionotropic, NMDA 1(RAT)
The Johns Hopkins University
Curated by PDSP Ki Database
The Johns Hopkins University
Curated by PDSP Ki Database
TargetGlutamate receptor ionotropic, NMDA 1(RAT)
The Johns Hopkins University
Curated by PDSP Ki Database
The Johns Hopkins University
Curated by PDSP Ki Database
TargetGlutamate receptor ionotropic, NMDA 1(RAT)
The Johns Hopkins University
Curated by PDSP Ki Database
The Johns Hopkins University
Curated by PDSP Ki Database
TargetGlutamate receptor ionotropic, NMDA 1(RAT)
The Johns Hopkins University
Curated by PDSP Ki Database
The Johns Hopkins University
Curated by PDSP Ki Database
TargetGlutamate receptor ionotropic, NMDA 1(RAT)
The Johns Hopkins University
Curated by PDSP Ki Database
The Johns Hopkins University
Curated by PDSP Ki Database
TargetGlutamate receptor ionotropic, NMDA 1(RAT)
The Johns Hopkins University
Curated by PDSP Ki Database
The Johns Hopkins University
Curated by PDSP Ki Database
TargetGlutamate receptor ionotropic, NMDA 1(RAT)
The Johns Hopkins University
Curated by PDSP Ki Database
The Johns Hopkins University
Curated by PDSP Ki Database
TargetGlutamate receptor ionotropic, NMDA 1(RAT)
The Johns Hopkins University
Curated by PDSP Ki Database
The Johns Hopkins University
Curated by PDSP Ki Database
TargetGlutamate receptor ionotropic, NMDA 1(RAT)
The Johns Hopkins University
Curated by PDSP Ki Database
The Johns Hopkins University
Curated by PDSP Ki Database
TargetGlutamate receptor ionotropic, NMDA 1(RAT)
The Johns Hopkins University
Curated by PDSP Ki Database
The Johns Hopkins University
Curated by PDSP Ki Database
TargetGlutamate receptor ionotropic, NMDA 1(RAT)
The Johns Hopkins University
Curated by PDSP Ki Database
The Johns Hopkins University
Curated by PDSP Ki Database
TargetGlutamate receptor ionotropic, NMDA 1(RAT)
The Johns Hopkins University
Curated by PDSP Ki Database
The Johns Hopkins University
Curated by PDSP Ki Database
TargetGlutamate receptor ionotropic, NMDA 1(RAT)
The Johns Hopkins University
Curated by PDSP Ki Database
The Johns Hopkins University
Curated by PDSP Ki Database
TargetGlutamate receptor ionotropic, NMDA 1(RAT)
The Johns Hopkins University
Curated by PDSP Ki Database
The Johns Hopkins University
Curated by PDSP Ki Database
TargetGlutamate receptor ionotropic, NMDA 1(RAT)
The Johns Hopkins University
Curated by PDSP Ki Database
The Johns Hopkins University
Curated by PDSP Ki Database
TargetGlutamate receptor ionotropic, NMDA 1(RAT)
The Johns Hopkins University
Curated by PDSP Ki Database
The Johns Hopkins University
Curated by PDSP Ki Database
Affinity DataKi: 93nMAssay Description:Displacement of Eu-H3/15 from human RXFP3 receptor expressed in CHOK1 cells by time-resolved fluorescent whole cell binding assayMore data for this Ligand-Target Pair
Affinity DataKi: 107nMAssay Description:Displacement of Eu-H3/15 from human RXFP3 receptor expressed in CHOK1 cells by time-resolved fluorescent whole cell binding assayMore data for this Ligand-Target Pair
Affinity DataKi: 110nMAssay Description:Displacement of europium-labelled H3 relaxin-B chain/INSL-5 chain from RXFP3 expressed in CHO-K1 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 110nMAssay Description:Displacement of Eu-H3/15 from human RXFP3 receptor expressed in CHOK1 cells by time-resolved fluorescent whole cell binding assayMore data for this Ligand-Target Pair
Affinity DataKi: 112nMAssay Description:Displacement of europium-labelled H3 relaxin-B chain/INSL-5 chain from RXFP3 expressed in CHO-K1 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 112nMAssay Description:Displacement of europium-labelled H3 relaxin-B chain/INSL-5 chain from RXFP3 expressed in CHO-K1 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 123nMAssay Description:Displacement of Eu-INSL5 from human RXFP4 receptor expressed in CHOK1 cells by time-resolved fluorescent whole cell binding assayMore data for this Ligand-Target Pair
Affinity DataKi: 151nMAssay Description:Displacement of Eu-H3/15 from human RXFP3 receptor expressed in CHOK1 cells by time-resolved fluorescent whole cell binding assayMore data for this Ligand-Target Pair
Affinity DataKi: 229nMAssay Description:Displacement of Eu-H3/15 from human RXFP3 receptor expressed in CHOK1 cells by time-resolved fluorescent whole cell binding assayMore data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 1(RAT)
The Johns Hopkins University
Curated by PDSP Ki Database
The Johns Hopkins University
Curated by PDSP Ki Database
Affinity DataKi: 275nMAssay Description:Displacement of Eu-INSL5 from human RXFP4 receptor expressed in CHOK1 cells by time-resolved fluorescent whole cell binding assayMore data for this Ligand-Target Pair
Affinity DataKi: 275nMAssay Description:Displacement of Eu-H3/15 from human RXFP3 receptor expressed in CHOK1 cells by time-resolved fluorescent whole cell binding assayMore data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 1(RAT)
The Johns Hopkins University
Curated by PDSP Ki Database
The Johns Hopkins University
Curated by PDSP Ki Database
TargetGlutamate receptor ionotropic, NMDA 1(RAT)
The Johns Hopkins University
Curated by PDSP Ki Database
The Johns Hopkins University
Curated by PDSP Ki Database


 3D Structure (crystal)
3D Structure (crystal)