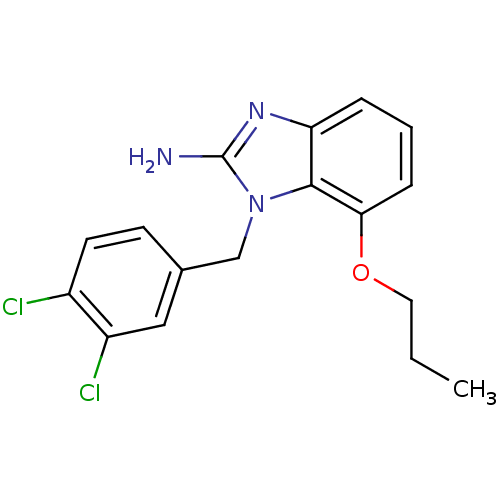
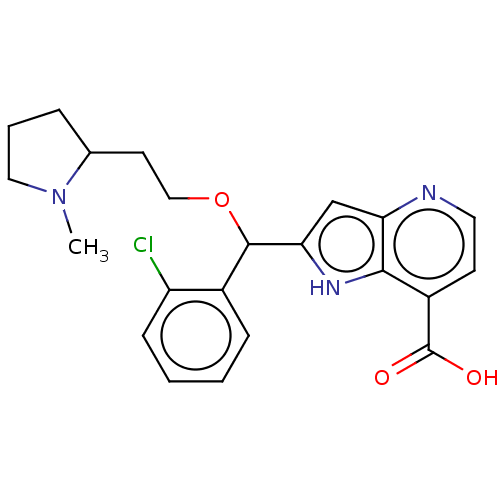
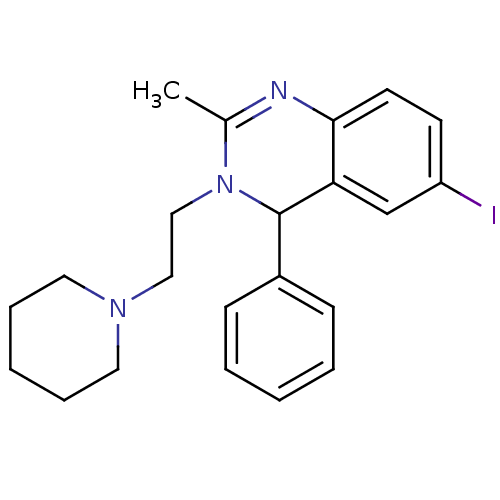
Affinity DataKi: 7nM ΔG°: -46.2kJ/molepH: 6.0 T: 2°CAssay Description:TbPTR1 activity was measured in 96-well microtiter plates via reduction of cytochrome c (cytc) as a result of the enzymatic production of tetrahydrob...More data for this Ligand-Target Pair
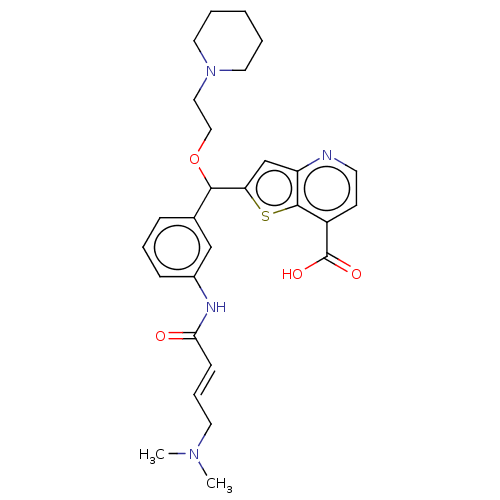
Affinity DataKi: 47nM ΔG°: -41.5kJ/molepH: 6.0 T: 2°CAssay Description:TbPTR1 activity was measured in 96-well microtiter plates via reduction of cytochrome c (cytc) as a result of the enzymatic production of tetrahydrob...More data for this Ligand-Target Pair
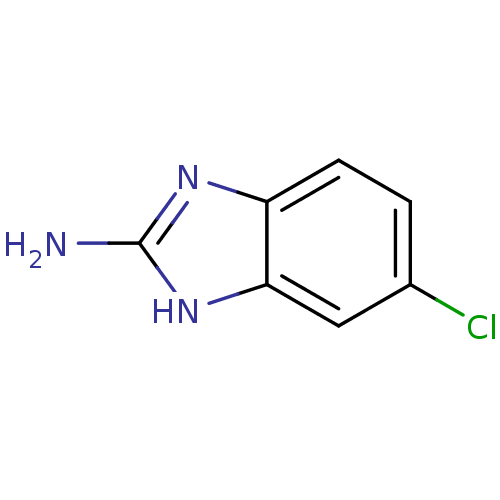
Affinity DataKi: 190nMAssay Description:Competitive inhibition of recombinant trypanothione reductase from Trypanosoma brucei brucei S427 by Lineweaver burk methodMore data for this Ligand-Target Pair
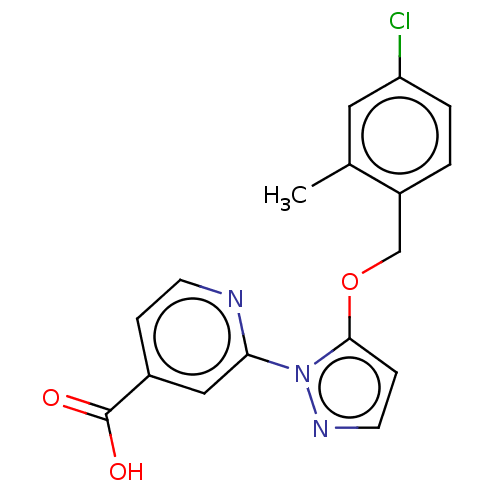
Affinity DataKi: 270nMAssay Description:Competitive inhibition of recombinant trypanothione reductase from Trypanosoma brucei brucei S427 by Lineweaver burk methodMore data for this Ligand-Target Pair
Affinity DataKi: 320nMAssay Description:Competitive inhibition of recombinant trypanothione reductase from Trypanosoma brucei brucei S427 by Lineweaver burk methodMore data for this Ligand-Target Pair
Affinity DataKi: 400nM ΔG°: -36.3kJ/molepH: 6.0 T: 2°CAssay Description:TbPTR1 activity was measured in 96-well microtiter plates via reduction of cytochrome c (cytc) as a result of the enzymatic production of tetrahydrob...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University Of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University Of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataKi: 410nMAssay Description:Irreversible inhibition of KDM5A ARID/PhD1 domain deletion mutant (1 to 739 residues) (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 440nMAssay Description:Competitive inhibition of recombinant trypanothione reductase from Trypanosoma brucei brucei S427 by Lineweaver burk methodMore data for this Ligand-Target Pair
Affinity DataKi: 510nM ΔG°: -35.7kJ/molepH: 6.0 T: 2°CAssay Description:TbPTR1 activity was measured in 96-well microtiter plates via reduction of cytochrome c (cytc) as a result of the enzymatic production of tetrahydrob...More data for this Ligand-Target Pair
Affinity DataKi: 1.46E+3nMAssay Description:Mixed type inhibition of recombinant trypanothione reductase from Trypanosoma brucei brucei S427 by Lineweaver burk methodMore data for this Ligand-Target Pair
Affinity DataKi: 1.78E+3nMAssay Description:Mixed type inhibition of recombinant trypanothione reductase from Trypanosoma brucei brucei S427 by Lineweaver burk methodMore data for this Ligand-Target Pair
Affinity DataKi: 2.27E+3nMAssay Description:Mixed type inhibition of recombinant trypanothione reductase from Trypanosoma brucei brucei S427 by Lineweaver burk methodMore data for this Ligand-Target Pair
Affinity DataKi: 9.80E+3nM ΔG°: -28.4kJ/molepH: 6.0 T: 2°CAssay Description:TbPTR1 activity was measured in 96-well microtiter plates via reduction of cytochrome c (cytc) as a result of the enzymatic production of tetrahydrob...More data for this Ligand-Target Pair
Affinity DataKi: 1.06E+4nM ΔG°: -28.2kJ/molepH: 6.0 T: 2°CAssay Description:TbPTR1 activity was measured in 96-well microtiter plates via reduction of cytochrome c (cytc) as a result of the enzymatic production of tetrahydrob...More data for this Ligand-Target Pair
Affinity DataKi: 2.11E+4nM ΔG°: -26.5kJ/molepH: 6.0 T: 2°CAssay Description:TbPTR1 activity was measured in 96-well microtiter plates via reduction of cytochrome c (cytc) as a result of the enzymatic production of tetrahydrob...More data for this Ligand-Target Pair
Affinity DataKi: 2.39E+4nM ΔG°: -26.2kJ/molepH: 6.0 T: 2°CAssay Description:TbPTR1 activity was measured in 96-well microtiter plates via reduction of cytochrome c (cytc) as a result of the enzymatic production of tetrahydrob...More data for this Ligand-Target Pair
Affinity DataKi: 1.41E+5nM ΔG°: -21.8kJ/molepH: 6.0 T: 2°CAssay Description:TbPTR1 activity was measured in 96-well microtiter plates via reduction of cytochrome c (cytc) as a result of the enzymatic production of tetrahydrob...More data for this Ligand-Target Pair
Affinity DataKi: >2.00E+5nM ΔG°: >-21.0kJ/molepH: 6.0 T: 2°CAssay Description:TbPTR1 activity was measured in 96-well microtiter plates via reduction of cytochrome c (cytc) as a result of the enzymatic production of tetrahydrob...More data for this Ligand-Target Pair
Affinity DataKi: 2.88E+5nM ΔG°: -20.1kJ/molepH: 6.0 T: 2°CAssay Description:TbPTR1 activity was measured in 96-well microtiter plates via reduction of cytochrome c (cytc) as a result of the enzymatic production of tetrahydrob...More data for this Ligand-Target Pair
Affinity DataIC50: 25nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
Affinity DataIC50: 30nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
Affinity DataIC50: 40nMAssay Description:Demethylase reactions and AlphaScreen assays were performed as described (Sayegh et al., 2013), with a few exceptions. All demethylase buffers contai...More data for this Ligand-Target Pair
Affinity DataIC50: 59nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University Of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University Of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 60nMAssay Description:Irreversible inhibition of 0.4 uM KDM5A ARID/PhD1 domain deletion mutant (1 to 739 residues) (unknown origin) using 100 uM alpha-KG by succinate-glo ...More data for this Ligand-Target Pair
Affinity DataIC50: 61nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
Affinity DataIC50: 67nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
Affinity DataIC50: 74nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University Of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University Of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 100nMAssay Description:Irreversible inhibition of KDM5A ARID/PhD1 domain deletion mutant (1 to 739 residues) (unknown origin) using 10 uM alpha-KG by succinate-glo demethyl...More data for this Ligand-Target Pair
Affinity DataIC50: 100nMAssay Description:Demethylase reactions and AlphaScreen assays were performed as described (Sayegh et al., 2013), with a few exceptions. All demethylase buffers contai...More data for this Ligand-Target Pair
Affinity DataIC50: 110nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University Of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University Of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 110nMAssay Description:Irreversible inhibition of KDM5A ARID/PhD1 domain deletion mutant (1 to 739 residues) (unknown origin) using alpha-KG measured immediately by succina...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University Of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University Of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 110nMAssay Description:Irreversible inhibition of KDM5A ARID/PhD1 domain deletion mutant (1 to 739 residues) (unknown origin) using 100 uM alpha-KG by succinate-glo demethy...More data for this Ligand-Target Pair
Affinity DataIC50: 120nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
Affinity DataIC50: 129nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
Affinity DataIC50: 160nMAssay Description:Demethylase reactions and AlphaScreen assays were performed as described (Sayegh et al., 2013), with a few exceptions. All demethylase buffers contai...More data for this Ligand-Target Pair
Affinity DataIC50: 175nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
Affinity DataIC50: 177nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
Affinity DataIC50: 180nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University Of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University Of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 180nMAssay Description:Irreversible inhibition of 0.8 uM KDM5A ARID/PhD1 domain deletion mutant (1 to 739 residues) (unknown origin) using 100 uM alpha-KG by succinate-glo ...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University Of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University Of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 180nMAssay Description:Irreversible inhibition of KDM5A ARID/PhD1 domain deletion mutant (1 to 739 residues) (unknown origin) using 1000 uM alpha-KG by succinate-glo demeth...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University Of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University Of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 220nMAssay Description:Irreversible inhibition of recombinant KDM5B ARID/PhD1 domain deletion mutant (1 to 755 residues) (unknown origin) expressed in Escherichia coli usin...More data for this Ligand-Target Pair
Affinity DataIC50: 230nMAssay Description:Inhibition of recombinant trypanothione reductase from Trypanosoma brucei brucei S427 by DTNB-coupled spectrophotometric assayMore data for this Ligand-Target Pair
Affinity DataIC50: 240nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University Of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University Of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 290nMAssay Description:Inhibition of recombinant human C-terminal FLAG-tagged KDM5A (1 to 1090 residues) expressed in baculovirus infected sf9 cells using biotinylated H3K4...More data for this Ligand-Target Pair
Affinity DataIC50: 300nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
Affinity DataIC50: 303nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University Of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University Of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 320nMAssay Description:Irreversible inhibition of recombinant KDM5B ARID/PhD1 domain deletion mutant (1 to 755 residues) (unknown origin) expressed in Escherichia coli usin...More data for this Ligand-Target Pair
Affinity DataIC50: 330nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
Affinity DataIC50: 350nMAssay Description:Inhibition of recombinant trypanothione reductase from Trypanosoma brucei brucei S427 by DTNB-coupled spectrophotometric assayMore data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University Of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University Of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 360nMAssay Description:Irreversible inhibition of 1.6 uM KDM5A ARID/PhD1 domain deletion mutant (1 to 739 residues) (unknown origin) using 100 uM alpha-KG by succinate-glo ...More data for this Ligand-Target Pair


 3D Structure (crystal)
3D Structure (crystal)