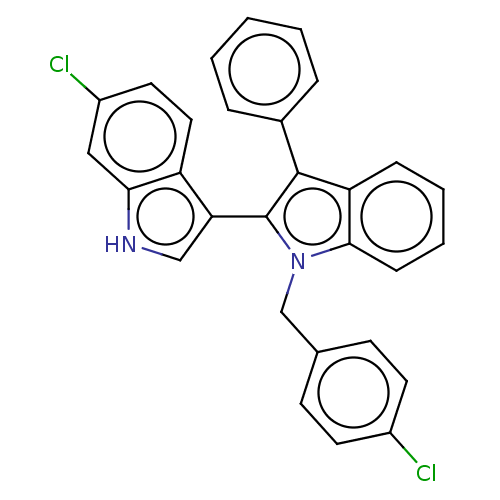
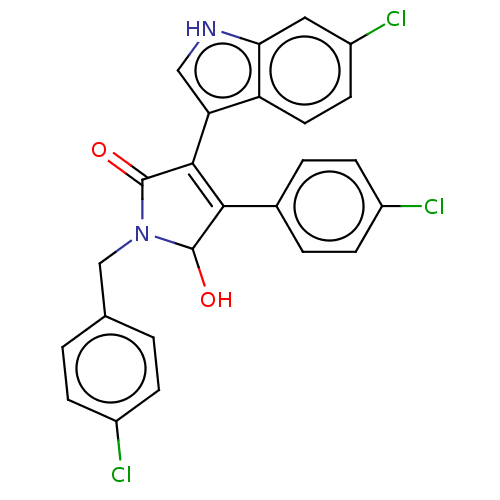
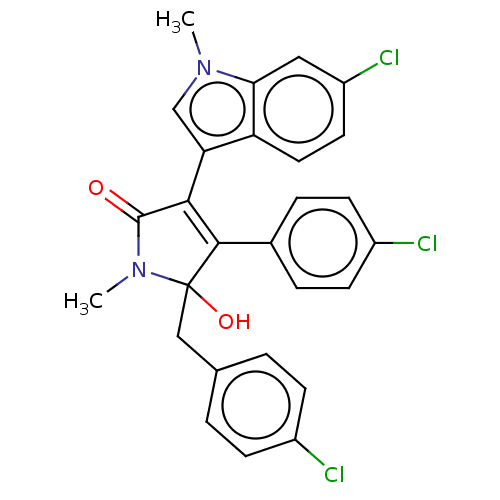
Affinity DataKi: 200nMAssay Description:Competition binding affinity to MDMX (unknown origin) using p53 mimicking peptide TSFAEYWNLLSP after 30 mins by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetE3 ubiquitin-protein ligase Mdm2(Homo sapiens (Human))
University Of Groningen
Curated by ChEMBL
University Of Groningen
Curated by ChEMBL
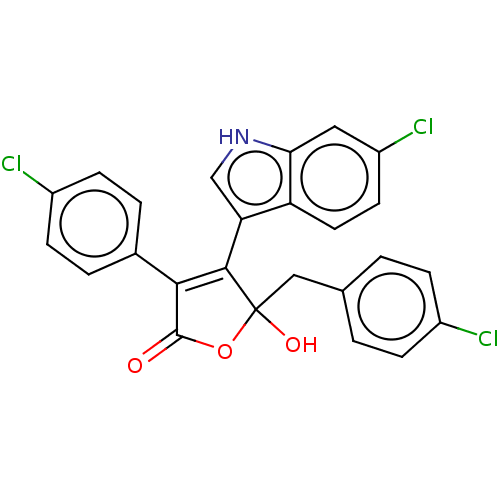
Affinity DataKi: 700nMAssay Description:Competition binding affinity to MDM2 (unknown origin) using p53 mimicking peptide TSFAEYWNLLSP after 30 mins by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetE3 ubiquitin-protein ligase Mdm2(Homo sapiens (Human))
University Of Groningen
Curated by ChEMBL
University Of Groningen
Curated by ChEMBL
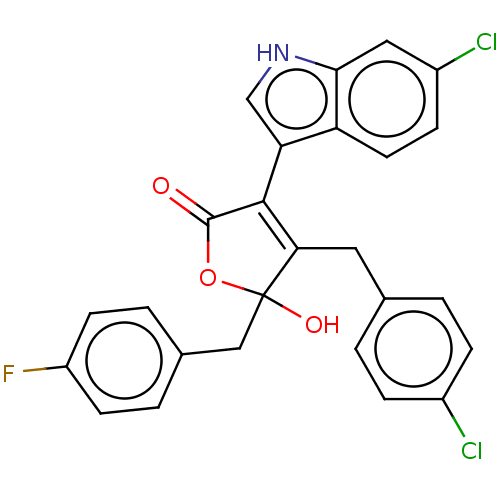
Affinity DataKi: 920nMAssay Description:Competition binding affinity to MDM2 (unknown origin) using p53 mimicking peptide TSFAEYWNLLSP after 30 mins by fluorescence polarization assayMore data for this Ligand-Target Pair
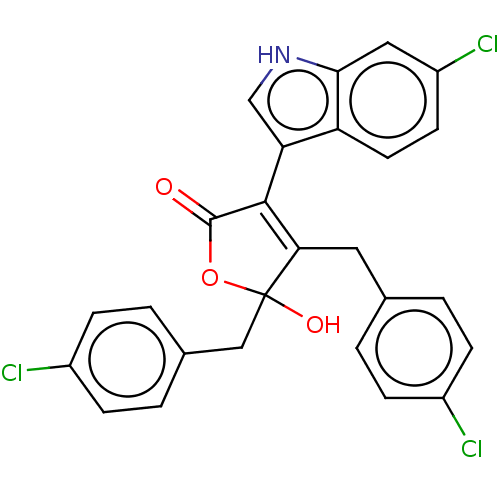
Affinity DataKi: 1.50E+3nMAssay Description:Competition binding affinity to MDMX (unknown origin) using p53 mimicking peptide TSFAEYWNLLSP after 30 mins by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetE3 ubiquitin-protein ligase Mdm2(Homo sapiens (Human))
University Of Groningen
Curated by ChEMBL
University Of Groningen
Curated by ChEMBL
Affinity DataKi: 1.80E+3nMAssay Description:Competition binding affinity to MDM2 (unknown origin) using p53 mimicking peptide TSFAEYWNLLSP after 30 mins by fluorescence polarization assayMore data for this Ligand-Target Pair
Affinity DataKi: 5.00E+3nMAssay Description:Competition binding affinity to MDMX (unknown origin) using p53 mimicking peptide TSFAEYWNLLSP after 30 mins by fluorescence polarization assayMore data for this Ligand-Target Pair
Affinity DataKi: 6.09E+3nMAssay Description:Competition binding affinity to MDMX (unknown origin) using p53 mimicking peptide TSFAEYWNLLSP after 30 mins by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetE3 ubiquitin-protein ligase Mdm2(Homo sapiens (Human))
University Of Groningen
Curated by ChEMBL
University Of Groningen
Curated by ChEMBL
Affinity DataKi: 6.70E+3nMAssay Description:Competition binding affinity to MDM2 (unknown origin) using p53 mimicking peptide TSFAEYWNLLSP after 30 mins by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetE3 ubiquitin-protein ligase Mdm2(Homo sapiens (Human))
University Of Groningen
Curated by ChEMBL
University Of Groningen
Curated by ChEMBL
Affinity DataKi: 1.00E+4nMAssay Description:Competition binding affinity to MDM2 (unknown origin) using p53 mimicking peptide TSFAEYWNLLSP after 30 mins by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetE3 ubiquitin-protein ligase Mdm2(Homo sapiens (Human))
University Of Groningen
Curated by ChEMBL
University Of Groningen
Curated by ChEMBL
Affinity DataKi: 1.70E+4nMAssay Description:Competition binding affinity to MDM2 (unknown origin) using p53 mimicking peptide TSFAEYWNLLSP after 30 mins by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetE3 ubiquitin-protein ligase Mdm2(Homo sapiens (Human))
University Of Groningen
Curated by ChEMBL
University Of Groningen
Curated by ChEMBL
Affinity DataKi: >2.20E+4nMAssay Description:Competition binding affinity to MDM2 (unknown origin) using p53 mimicking peptide TSFAEYWNLLSP after 30 mins by fluorescence polarization assayMore data for this Ligand-Target Pair
Affinity DataKi: 3.60E+4nMAssay Description:Competition binding affinity to MDMX (unknown origin) using p53 mimicking peptide TSFAEYWNLLSP after 30 mins by fluorescence polarization assayMore data for this Ligand-Target Pair
Affinity DataKi: >6.00E+4nMAssay Description:Competition binding affinity to MDMX (unknown origin) using p53 mimicking peptide TSFAEYWNLLSP after 30 mins by fluorescence polarization assayMore data for this Ligand-Target Pair
Affinity DataKi: >6.00E+4nMAssay Description:Competition binding affinity to MDMX (unknown origin) using p53 mimicking peptide TSFAEYWNLLSP after 30 mins by fluorescence polarization assayMore data for this Ligand-Target Pair
Affinity DataKi: >6.00E+4nMAssay Description:Competition binding affinity to MDMX (unknown origin) using p53 mimicking peptide TSFAEYWNLLSP after 30 mins by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetE3 ubiquitin-protein ligase Mdm2(Homo sapiens (Human))
University Of Groningen
Curated by ChEMBL
University Of Groningen
Curated by ChEMBL
Affinity DataKi: >6.00E+4nMAssay Description:Competition binding affinity to MDM2 (unknown origin) using p53 mimicking peptide TSFAEYWNLLSP after 30 mins by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetProgrammed cell death 1 ligand 1(Homo sapiens (Human))
Jagiellonian University
Curated by ChEMBL
Jagiellonian University
Curated by ChEMBL
Affinity DataIC50: 1.40nMAssay Description:Binding affinity to His-tagged PD-L1 (unknown origin) assessed as reduction in PD-L1/PD1-Ig interaction preincubated with PD-L1 for 15 mins followed ...More data for this Ligand-Target Pair
TargetProgrammed cell death 1 ligand 1(Homo sapiens (Human))
Jagiellonian University
Curated by ChEMBL
Jagiellonian University
Curated by ChEMBL
Affinity DataIC50: 2.30nMAssay Description:Binding affinity to His-tagged PD-L1 (unknown origin) assessed as reduction in PD-L1/PD1-Ig interaction preincubated with PD-L1 for 15 mins followed ...More data for this Ligand-Target Pair
TargetProgrammed cell death 1 ligand 1(Homo sapiens (Human))
Jagiellonian University
Curated by ChEMBL
Jagiellonian University
Curated by ChEMBL
Affinity DataIC50: 6nMAssay Description:Binding affinity to His-tagged PD-L1 (unknown origin) assessed as reduction in PD-L1/PD1-Ig interaction preincubated with PD-L1 for 15 mins followed ...More data for this Ligand-Target Pair
TargetProgrammed cell death 1 ligand 1(Homo sapiens (Human))
Jagiellonian University
Curated by ChEMBL
Jagiellonian University
Curated by ChEMBL
Affinity DataIC50: 6nMAssay Description:Binding affinity to His-tagged PD-L1 (unknown origin) assessed as reduction in PD-L1/PD1-Ig interaction preincubated with PD-L1 for 15 mins followed ...More data for this Ligand-Target Pair
TargetProgrammed cell death 1 ligand 1(Homo sapiens (Human))
Jagiellonian University
Curated by ChEMBL
Jagiellonian University
Curated by ChEMBL
Affinity DataIC50: 18nMAssay Description:Binding affinity to His-tagged PD-L1 (unknown origin) assessed as reduction in PD-L1/PD1-Ig interaction preincubated with PD-L1 for 15 mins followed ...More data for this Ligand-Target Pair
TargetProgrammed cell death 1 ligand 1(Homo sapiens (Human))
Jagiellonian University
Curated by ChEMBL
Jagiellonian University
Curated by ChEMBL
Affinity DataIC50: 80nMAssay Description:Binding affinity to His-tagged PD-L1 (unknown origin) assessed as reduction in PD-L1/PD1-Ig interaction preincubated with PD-L1 for 15 mins followed ...More data for this Ligand-Target Pair
TargetProgrammed cell death 1 ligand 1(Homo sapiens (Human))
Jagiellonian University
Curated by ChEMBL
Jagiellonian University
Curated by ChEMBL
Affinity DataIC50: 146nMAssay Description:Binding affinity to His-tagged PD-L1 (unknown origin) assessed as reduction in PD-L1/PD1-Ig interaction preincubated with PD-L1 for 15 mins followed ...More data for this Ligand-Target Pair
Affinity DataKd: 1.30E+3nMpH: 7.4 T: 2°CAssay Description:MST (NanoTemper Technologies GmbH) was used to determine the binding affinities between Mdm2 (residues 1−118 T47W; 500 nM) and inhibitors. T47W...More data for this Ligand-Target Pair
Affinity DataKd: 7.00E+3nMpH: 7.4 T: 2°CAssay Description:MST (NanoTemper Technologies GmbH) was used to determine the binding affinities between Mdm2 (residues 1−118 T47W; 500 nM) and inhibitors. T47W...More data for this Ligand-Target Pair
TargetProgrammed cell death 1 ligand 1(Homo sapiens (Human))
Jagiellonian University
Curated by ChEMBL
Jagiellonian University
Curated by ChEMBL
Affinity DataKd: <1.00E+3nMAssay Description:Binding affinity to 15N-labeled human PD-L1 (18 to 134 residues) expressed in Escherichia coli BL21(DE3) by 1H-15N 2D HMQC NMR spectroscopic analysisMore data for this Ligand-Target Pair
TargetProgrammed cell death 1 ligand 1(Homo sapiens (Human))
Jagiellonian University
Curated by ChEMBL
Jagiellonian University
Curated by ChEMBL
Affinity DataKd: <1.00E+3nMAssay Description:Binding affinity to 15N-labeled human PD-L1 (18 to 134 residues) expressed in Escherichia coli BL21(DE3) by 1H-15N 2D HMQC NMR spectroscopic analysisMore data for this Ligand-Target Pair
Affinity DataKd: 2.60E+4nMpH: 7.4 T: 2°CAssay Description:MST (NanoTemper Technologies GmbH) was used to determine the binding affinities between Mdm2 (residues 1−118 T47W; 500 nM) and inhibitors. T47W...More data for this Ligand-Target Pair
Affinity DataKd: 2.87E+4nMpH: 7.4 T: 2°CAssay Description:MST (NanoTemper Technologies GmbH) was used to determine the binding affinities between Mdm2 (residues 1−118 T47W; 500 nM) and inhibitors. T47W...More data for this Ligand-Target Pair
Affinity DataKd: 1.50E+3nMpH: 7.4 T: 2°CAssay Description:MST (NanoTemper Technologies GmbH) was used to determine the binding affinities between Mdm2 (residues 1−118 T47W; 500 nM) and inhibitors. T47W...More data for this Ligand-Target Pair
Affinity DataKd: 3.10E+3nMpH: 7.4 T: 2°CAssay Description:MST (NanoTemper Technologies GmbH) was used to determine the binding affinities between Mdm2 (residues 1−118 T47W; 500 nM) and inhibitors. T47W...More data for this Ligand-Target Pair
Affinity DataKd: 2.10E+3nMpH: 7.4 T: 2°CAssay Description:MST (NanoTemper Technologies GmbH) was used to determine the binding affinities between Mdm2 (residues 1−118 T47W; 500 nM) and inhibitors. T47W...More data for this Ligand-Target Pair
Affinity DataKd: 2.44E+4nMpH: 7.4 T: 2°CAssay Description:MST (NanoTemper Technologies GmbH) was used to determine the binding affinities between Mdm2 (residues 1−118 T47W; 500 nM) and inhibitors. T47W...More data for this Ligand-Target Pair
Affinity DataKd: 3.04E+4nMpH: 7.4 T: 2°CAssay Description:MST (NanoTemper Technologies GmbH) was used to determine the binding affinities between Mdm2 (residues 1−118 T47W; 500 nM) and inhibitors. T47W...More data for this Ligand-Target Pair
Affinity DataKd: 4.05E+4nMpH: 7.4 T: 2°CAssay Description:MST (NanoTemper Technologies GmbH) was used to determine the binding affinities between Mdm2 (residues 1−118 T47W; 500 nM) and inhibitors. T47W...More data for this Ligand-Target Pair
Affinity DataKd: 2.77E+4nMpH: 7.4 T: 2°CAssay Description:MST (NanoTemper Technologies GmbH) was used to determine the binding affinities between Mdm2 (residues 1−118 T47W; 500 nM) and inhibitors. T47W...More data for this Ligand-Target Pair
Affinity DataKd: 1.87E+4nMpH: 7.4 T: 2°CAssay Description:MST (NanoTemper Technologies GmbH) was used to determine the binding affinities between Mdm2 (residues 1−118 T47W; 500 nM) and inhibitors. T47W...More data for this Ligand-Target Pair
Affinity DataKd: 500nMpH: 7.4 T: 2°CAssay Description:MST (NanoTemper Technologies GmbH) was used to determine the binding affinities between Mdm2 (residues 1−118 T47W; 500 nM) and inhibitors. T47W...More data for this Ligand-Target Pair
Affinity DataKd: 6.80E+4nMpH: 7.4 T: 2°CAssay Description:Uniform 15N isotope labeling was achieved by expression of the protein in the M9 minimal media containing 15NH4Cl as the sole nitrogen source. The fi...More data for this Ligand-Target Pair
Affinity DataKd: 6.00E+4nMpH: 7.4 T: 2°CAssay Description:Uniform 15N isotope labeling was achieved by expression of the protein in the M9 minimal media containing 15NH4Cl as the sole nitrogen source. The fi...More data for this Ligand-Target Pair
Affinity DataKd: 1.80E+4nMpH: 7.4 T: 2°CAssay Description:Uniform 15N isotope labeling was achieved by expression of the protein in the M9 minimal media containing 15NH4Cl as the sole nitrogen source. The fi...More data for this Ligand-Target Pair
Affinity DataKd: 8.60E+4nMpH: 7.4 T: 2°CAssay Description:Uniform 15N isotope labeling was achieved by expression of the protein in the M9 minimal media containing 15NH4Cl as the sole nitrogen source. The fi...More data for this Ligand-Target Pair
Affinity DataKd: <1.00E+3nMpH: 7.4 T: 2°CAssay Description:Uniform 15N isotope labeling was achieved by expression of the protein in the M9 minimal media containing 15NH4Cl as the sole nitrogen source. The fi...More data for this Ligand-Target Pair
Affinity DataKd: 4.00E+4nMpH: 7.4 T: 2°CAssay Description:Uniform 15N isotope labeling was achieved by expression of the protein in the M9 minimal media containing 15NH4Cl as the sole nitrogen source. The fi...More data for this Ligand-Target Pair
Affinity DataKd: 1.64E+5nMpH: 7.4 T: 2°CAssay Description:Uniform 15N isotope labeling was achieved by expression of the protein in the M9 minimal media containing 15NH4Cl as the sole nitrogen source. The fi...More data for this Ligand-Target Pair
Affinity DataKd: 2.46E+5nMpH: 7.4 T: 2°CAssay Description:Uniform 15N isotope labeling was achieved by expression of the protein in the M9 minimal media containing 15NH4Cl as the sole nitrogen source. The fi...More data for this Ligand-Target Pair
Affinity DataKd: 3.65E+5nMpH: 7.4 T: 2°CAssay Description:Uniform 15N isotope labeling was achieved by expression of the protein in the M9 minimal media containing 15NH4Cl as the sole nitrogen source. The fi...More data for this Ligand-Target Pair
Affinity DataKd: 1.92E+5nMpH: 7.4 T: 2°CAssay Description:Uniform 15N isotope labeling was achieved by expression of the protein in the M9 minimal media containing 15NH4Cl as the sole nitrogen source. The fi...More data for this Ligand-Target Pair
Affinity DataKd: 5.10E+4nMpH: 7.4 T: 2°CAssay Description:Uniform 15N isotope labeling was achieved by expression of the protein in the M9 minimal media containing 15NH4Cl as the sole nitrogen source. The fi...More data for this Ligand-Target Pair
Affinity DataKd: <1.00E+3nMpH: 7.4 T: 2°CAssay Description:Uniform 15N isotope labeling was achieved by expression of the protein in the M9 minimal media containing 15NH4Cl as the sole nitrogen source. The fi...More data for this Ligand-Target Pair




 3D Structure (crystal)
3D Structure (crystal)