Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Envelope glycoprotein gp160
Ligand
BDBM50252
Substrate
n/a
Meas. Tech.
SAR analysis of compounds that inhibit Human Immunodeficiency Virus Fusion.
IC50
5700±n/a nM
Citation
 PubChem, PC SAR analysis of compounds that inhibit Human Immunodeficiency Virus Fusion. PubChem Bioassay (2010)[AID]
PubChem, PC SAR analysis of compounds that inhibit Human Immunodeficiency Virus Fusion. PubChem Bioassay (2010)[AID] More Info.:
Target
Name:
Envelope glycoprotein gp160
Synonyms:
envelope glycoprotein
Type:
Enzyme Catalytic Domain
Mol. Mass.:
91798.58
Organism:
Human immunodeficiency virus 1
Description:
gi_45357394
Residue:
814
Sequence:
MRVKGIRRNYQHLWRWGTMLLGMLMICSAKEQLWVTAYYGVPVWKEATTTLFCASDAKAYDTEVHNVWATHACVPTDPNPREVVMGNVTEEFNIWNNSMVEQMHEDIISLWDESLKPCVKLTPLCVTFNCTNYNGTRNGTTTEPPEVKNCTTKETGIKNCSFNIATSGVEDRFKKEYALLYTADIVQIDNSSINYTLIGCNTSVITQACPKVSFEPIPIHYCAPAGFAILKCNNKTFNGKGPCTNVSTVQCTHGIRPVVSTQLLLNGSLAEEVVIRSDNFSDNAKTIIVQLKDPVVINCTRPNNNTRKGIRIGPGRTFYTTERIIGDIRQAHCNISRTQWNNTLRLIAAKLKKQFNNKTIIFRNSSGGDPEIVMHSFNCGGEFFYCNTTQLFNSTWVHNNTWVHNNTGNDTEEGTITLPCRIKQIINMWQEVGKAMYAPPIKGQIRCSSNITGLILTRDGGNTSSNNETFRPGGGDMRDNWRSELYKYKVVKIEPLGVAPTKARRRVVQREKRAVGMLGAMFLGFLGAAGSTMGAASLALTVQARQVVSGIVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARVLAVERYLGDQQLLGIWGCSGKLICTTAVPWNDSWSNKSLKYIWDNMTWMQWEKEIDIHVDTIYQLLEASQYQQEKNEKDLLELDKWESLWNWFDITNWLWYIKIFIMIVGGLIGLRIVFTVLSIVNRVRQGYSPLSLQTRLPTQRGPDRPEGIEEEGGERQRHIRSISEWILNNYPGRPAEPVPLQLPPLERLTLDCNEDCGTSGTQGVGSPQVFVESPPVLESGTKEECC
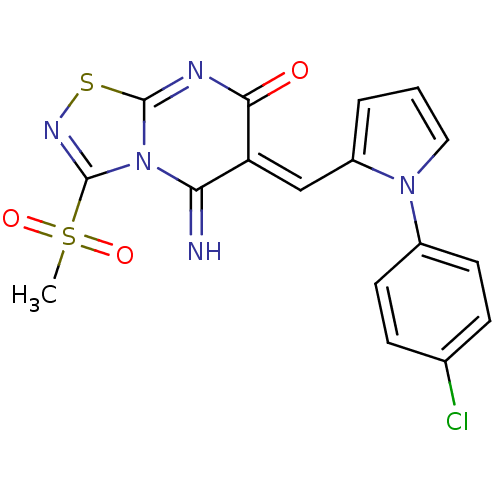
Inhibitor
Name:
BDBM50252
Synonyms:
(6Z)-5-azanylidene-6-[[1-(4-chlorophenyl)pyrrol-2-yl]methylidene]-3-methylsulfonyl-[1,2,4]thiadiazolo[4,5-a]pyrimidin-7-one | (6Z)-6-[[1-(4-chlorophenyl)-2-pyrrolyl]methylidene]-5-imino-3-methylsulfonyl-[1,2,4]thiadiazolo[4,5-a]pyrimidin-7-one | (6Z)-6-[[1-(4-chlorophenyl)pyrrol-2-yl]methylene]-5-imino-3-mesyl-[1,2,4]thiadiazolo[4,5-a]pyrimidin-7-one | (6Z)-6-[[1-(4-chlorophenyl)pyrrol-2-yl]methylidene]-5-imino-3-methylsulfonyl-[1,2,4]thiadiazolo[4,5-a]pyrimidin-7-one | (6Z)-6-{[1-(4-chlorophenyl)-1H-pyrrol-2-yl]methylene}-5-imino-3-(methylsulfonyl)-5,6-dihydro-7H-[1,2,4]thiadiazolo[4,5-a]pyrimidin-7-one | MLS000583161 | SMR000206347 | cid_12005346
Type:
Small organic molecule
Emp. Form.:
C17H12ClN5O3S2
Mol. Mass.:
433.892
SMILES:
CS(=O)(=O)C1=NSC2=NC(=O)\C(=C/c3cccn3-c3ccc(Cl)cc3)C(=N)N12 |t:4,7|