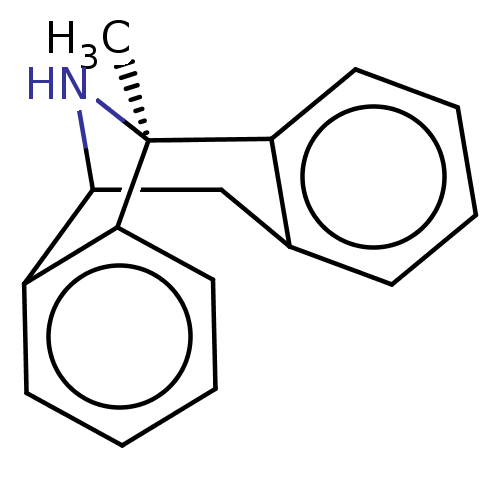
BDBM50000663 (+)-1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene::(+)-MK-801::(+)MK-801::(+/-) MK-8011-methyl-(9R,1R)-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene::(+/-)-1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene::(+/-)-MK801::(-)-1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene::(-)-MK801::(1S,9R)-1-methyl-16-azatetracyclo[7.6.1.0^{2,7}.0^{10,15}]hexadeca-2(7),3,5,10(15),11,13-hexaene::(1S,9R)-1-methyl-16-azatetracyclo[7.6.1.0^{2,7}.0^{10,15}]hexadeca-2,4,6,10(15),11,13-hexaene::(5S,10R)-(+)-5-methyl-10,11-dihydro-5Hdibenzo[a,d]cyclohepten-5,10-imine::(5S,10S)-(+)-5-methyl-10,11-dihydro-5Hdibenzo[a,d]cyclohepten-5,10-imine::(Dizocilpine)1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene::(MK-801)1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene::1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene::1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene(MK-801)::1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10,12,14-hexaene::1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2,4,6,10(15),11,13-hexaene::10,11-Dihydro-5-methyl-5H-dibenzo[a,d]cyclohepten-5,10-imine.(MK-801)::CHEMBL284237::MK-801::MK-801 (Dizocilpine)::MK-801,(+)::MK-801,(-)::dizocilpine
SMILES C[C@]12NC(Cc3ccccc13)c1ccccc21
InChI Key InChIKey=LBOJYSIDWZQNJS-LYKKTTPLSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 17 hits for monomerid = 50000663
Found 17 hits for monomerid = 50000663
National Institute Of Diabetes And Digestive And Kidney Diseases
Curated by ChEMBL
National Institute Of Diabetes And Digestive And Kidney Diseases
Curated by ChEMBL
National Institute Of Diabetes And Digestive And Kidney Diseases
Curated by ChEMBL
National Institute Of Diabetes And Digestive And Kidney Diseases
Curated by ChEMBL
Medical University Of Lublin
Curated by ChEMBL