Structure and Dynamics of Adrenomedullin Receptors AM1and AM2Reveal Key Mechanisms in the Control of Receptor Phenotype by Receptor Activity-Modifying Proteins.
Liang, Y.L., Belousoff, M.J., Fletcher, M.M., Zhang, X., Khoshouei, M., Deganutti, G., Koole, C., Furness, S.G.B., Miller, L.J., Hay, D.L., Christopoulos, A., Reynolds, C.A., Danev, R., Wootten, D., Sexton, P.M.(2020) Acs Pharmacol Transl Sci 3: 263-284
- PubMed: 32296767
- DOI: https://doi.org/10.1021/acsptsci.9b00080
- Primary Citation of Related Structures:
6UUN, 6UUS, 6UVA - PubMed Abstract:
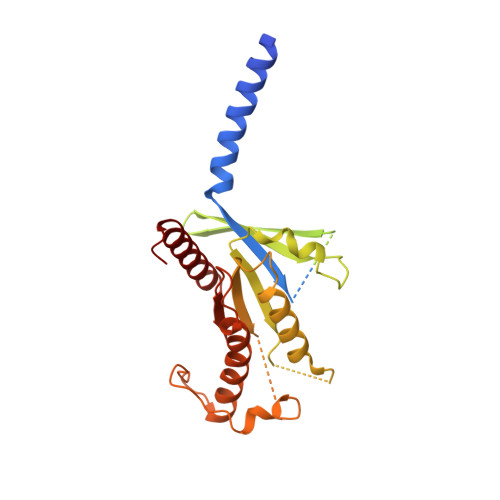
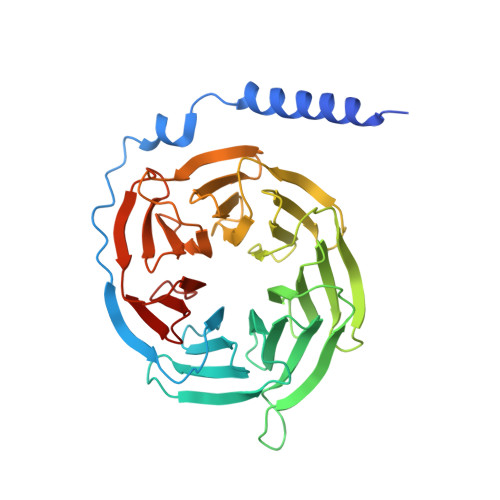
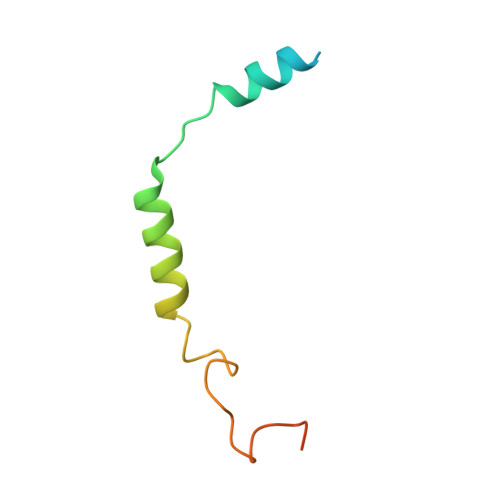
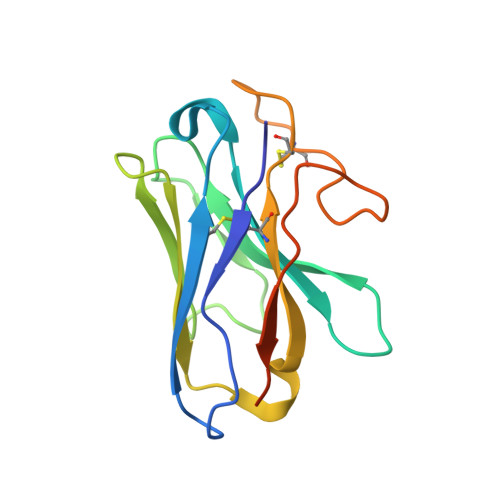
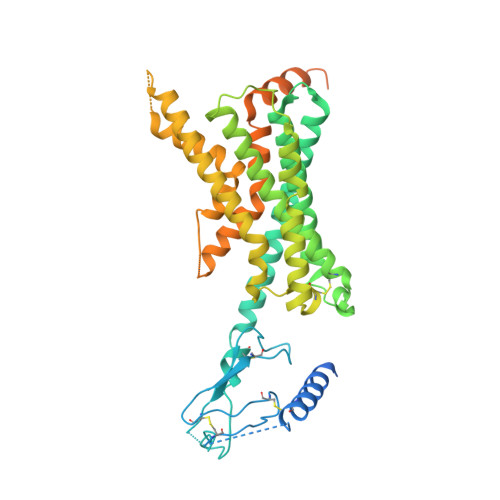
Adrenomedullin (AM) and calcitonin gene-related peptide (CGRP) receptors are critically important for metabolism, vascular tone, and inflammatory response. AM receptors are also required for normal lymphatic and blood vascular development and angiogenesis. They play a pivotal role in embryo implantation and fertility and can provide protection against hypoxic and oxidative stress. CGRP and AM receptors are heterodimers of the calcitonin receptor-like receptor (CLR) and receptor activity-modifying protein 1 (RAMP1) (CGRPR), as well as RAMP2 or RAMP3 (AM 1 R and AM 2 R, respectively). However, the mechanistic basis for RAMP modulation of CLR phenotype is unclear. In this study, we report the cryo-EM structure of the AM 1 R in complex with AM and Gs at a global resolution of 3.0 Å, and structures of the AM 2 R in complex with either AM or intermedin/adrenomedullin 2 (AM2) and Gs at 2.4 and 2.3 Å, respectively. The structures reveal distinctions in the primary orientation of the extracellular domains (ECDs) relative to the receptor core and distinct positioning of extracellular loop 3 (ECL3) that are receptor-dependent. Analysis of dynamic data present in the cryo-EM micrographs revealed additional distinctions in the extent of mobility of the ECDs. Chimeric exchange of the linker region of the RAMPs connecting the TM helix and the ECD supports a role for this segment in controlling receptor phenotype. Moreover, a subset of the motions of the ECD appeared coordinated with motions of the G protein relative to the receptor core, suggesting that receptor ECD dynamics could influence G protein interactions. This work provides fundamental advances in our understanding of GPCR function and how this can be allosterically modulated by accessory proteins.
Organizational Affiliation:
Drug Discovery Biology, Monash Institute of Pharmaceutical Sciences, Monash University, Parkville 3052, Victoria, Australia.