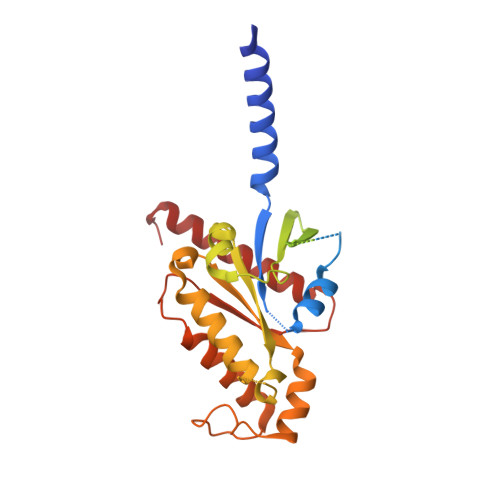
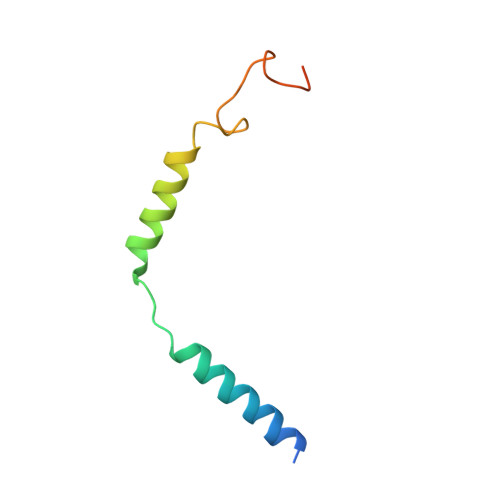
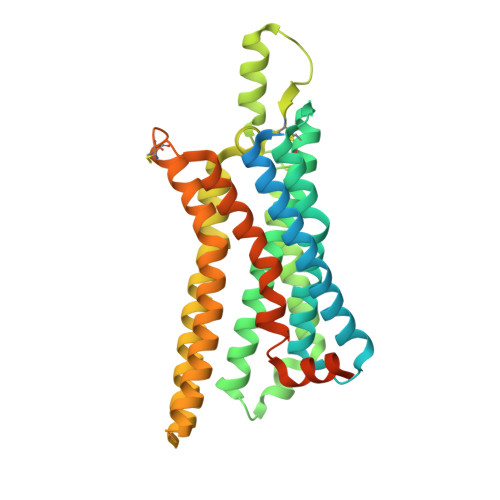
Structure of the adenosine-bound human adenosine A1receptor-Gicomplex.
Draper-Joyce, C.J., Khoshouei, M., Thal, D.M., Liang, Y.L., Nguyen, A.T.N., Furness, S.G.B., Venugopal, H., Baltos, J.A., Plitzko, J.M., Danev, R., Baumeister, W., May, L.T., Wootten, D., Sexton, P.M., Glukhova, A., Christopoulos, A.(2018) Nature 558: 559-563
- PubMed: 29925945
- DOI: https://doi.org/10.1038/s41586-018-0236-6
- Primary Citation of Related Structures:
6D9H - PubMed Abstract:
The class A adenosine A 1 receptor (A 1 R) is a G-protein-coupled receptor that preferentially couples to inhibitory G i/o heterotrimeric G proteins, has been implicated in numerous diseases, yet remains poorly targeted. Here we report the 3.6 Å structure of the human A 1 R in complex with adenosine and heterotrimeric G i2 protein determined by Volta phase plate cryo-electron microscopy. Compared to inactive A 1 R, there is contraction at the extracellular surface in the orthosteric binding site mediated via movement of transmembrane domains 1 and 2. At the intracellular surface, the G protein engages the A 1 R primarily via amino acids in the C terminus of the Gα i α5-helix, concomitant with a 10.5 Å outward movement of the A 1 R transmembrane domain 6. Comparison with the agonist-bound β 2 adrenergic receptor-G s -protein complex reveals distinct orientations for each G-protein subtype upon engagement with its receptor. This active A 1 R structure provides molecular insights into receptor and G-protein selectivity.
Organizational Affiliation:
Drug Discovery Biology and Department of Pharmacology, Monash Institute of Pharmaceutical Sciences, Monash University, Parkville, Victoria, Australia.