Identification of a non-competitive inhibitor of Plasmodium falciparum aspartate transcarbamoylase.
Lunev, S., Bosch, S.S., Batista, F.A., Wang, C., Li, J., Linzke, M., Kruithof, P., Chamoun, G., Domling, A.S.S., Wrenger, C., Groves, M.R.(2018) Biochem Biophys Res Commun 497: 835-842
- PubMed: 29476738
- DOI: https://doi.org/10.1016/j.bbrc.2018.02.112
- Primary Citation of Related Structures:
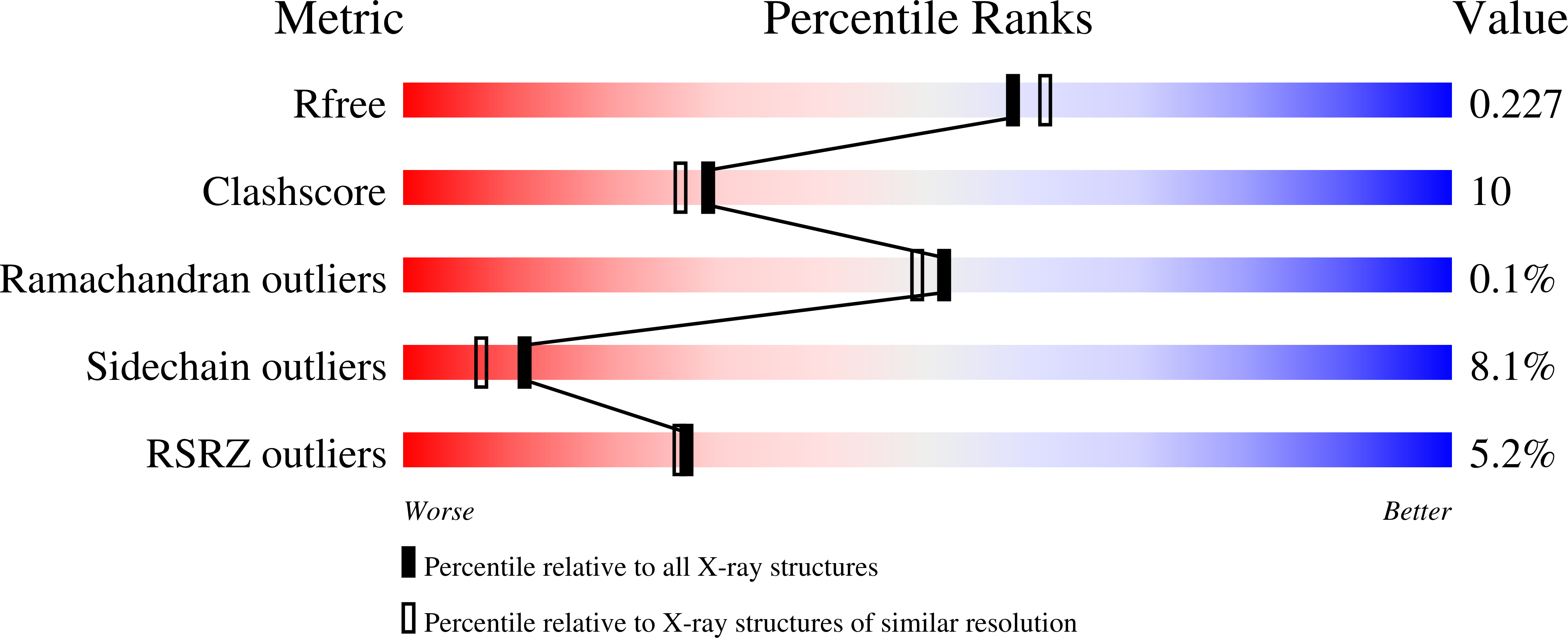
6FBA - PubMed Abstract:
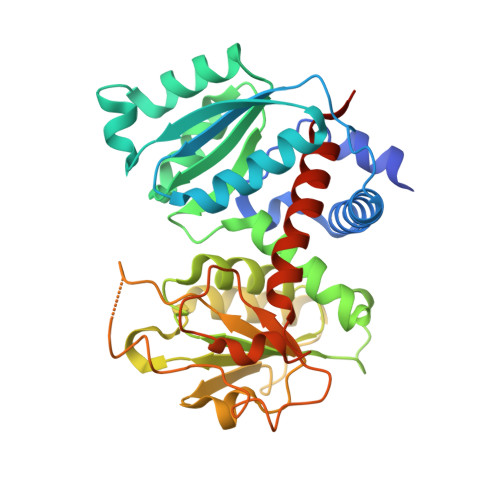
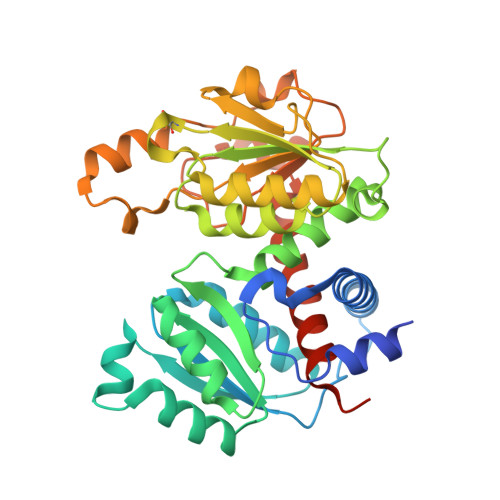
Aspartate transcarbamoylase catalyzes the second step of de-novo pyrimidine biosynthesis. As malarial parasites lack pyrimidine salvage machinery and rely on de-novo production for growth and proliferation, this pathway is a target for drug discovery. Previously, an apo crystal structure of aspartate transcarbamoylase from Plasmodium falciparum (PfATC) in its T-state has been reported. Here we present crystal structures of PfATC in the liganded R-state as well as in complex with the novel inhibitor, 2,3-napthalenediol, identified by high-throughput screening. Our data shows that 2,3-napthalediol binds in close proximity to the active site, implying an allosteric mechanism of inhibition. Furthermore, we report biophysical characterization of 2,3-napthalenediol. These data provide a promising starting point for structure based drug design targeting PfATC and malarial de-novo pyrimidine biosynthesis.
Organizational Affiliation:
Faculty of Science and Engineering, University of Groningen, Antonius Deusinglaan 1, Groningen, 9713AV, The Netherlands.