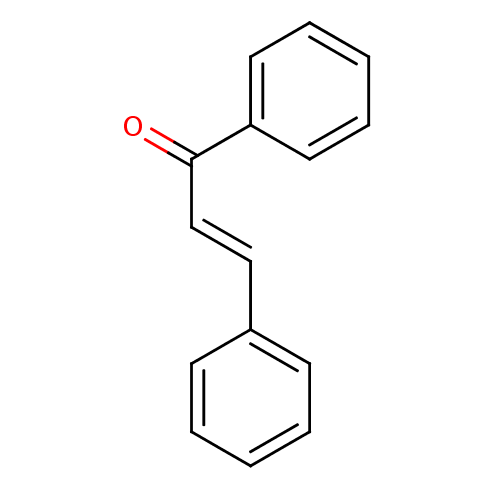
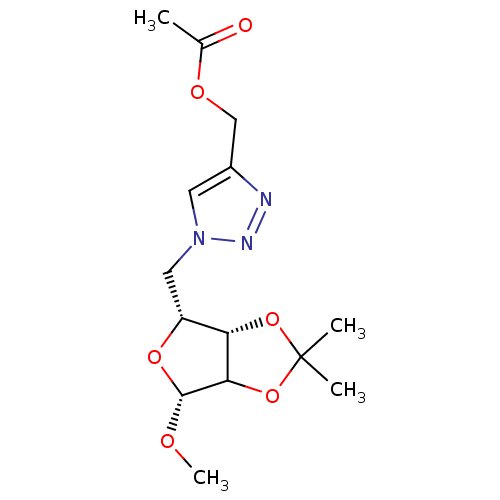
Affinity DataKi: 1.17E+4nMAssay Description:Non-competitive inhibition of porcine pancreatic alpha-amylase by Lineweaver-Burk plot analysisMore data for this Ligand-Target Pair
Affinity DataKi: 3.36E+4nM ΔG°: -6.34kcal/mole IC50: 3.58E+4nMpH: 7.0 T: 2°CAssay Description:To test whether GCTs share a common binding pocket on PPA with acarbose, we performed an experiment according to the method of Yonetani and Theorell.More data for this Ligand-Target Pair
Affinity DataKi: 4.80E+4nMAssay Description:Inhibition of porcine pancreatic alpha-amylase using soluble starch as substrate after 30 mins by Bernfeld methodMore data for this Ligand-Target Pair
Affinity DataKi: 2.04E+5nM ΔG°: -5.23kcal/mole IC50: 8.89E+4nMpH: 7.0 T: 2°CAssay Description:To test whether GCTs share a common binding pocket on PPA with acarbose, we performed an experiment according to the method of Yonetani and Theorell.More data for this Ligand-Target Pair
Affinity DataKi: 2.34E+5nM ΔG°: -5.15kcal/mole IC50: 1.25E+5nMpH: 7.0 T: 2°CAssay Description:To test whether GCTs share a common binding pocket on PPA with acarbose, we performed an experiment according to the method of Yonetani and Theorell.More data for this Ligand-Target Pair
Affinity DataKi: 3.42E+5nM ΔG°: -4.92kcal/mole IC50: 1.50E+5nMpH: 7.0 T: 2°CAssay Description:To test whether GCTs share a common binding pocket on PPA with acarbose, we performed an experiment according to the method of Yonetani and Theorell.More data for this Ligand-Target Pair
Affinity DataKi: 7.80E+5nM ΔG°: -4.41kcal/mole IC50: 3.61E+5nMpH: 7.0 T: 2°CAssay Description:To test whether GCTs share a common binding pocket on PPA with acarbose, we performed an experiment according to the method of Yonetani and Theorell.More data for this Ligand-Target Pair
Affinity DataKi: 1.01E+6nM ΔG°: -4.25kcal/mole IC50: 3.56E+5nMpH: 7.0 T: 2°CAssay Description:To test whether GCTs share a common binding pocket on PPA with acarbose, we performed an experiment according to the method of Yonetani and Theorell.More data for this Ligand-Target Pair
Affinity DataKi: <1.00E+7nMAssay Description:Inhibition of porcine pancreatic alpha-amylase using DP17 amylose as substrate preincubated for 5 mins followed by substrate addition by copper-bicin...More data for this Ligand-Target Pair