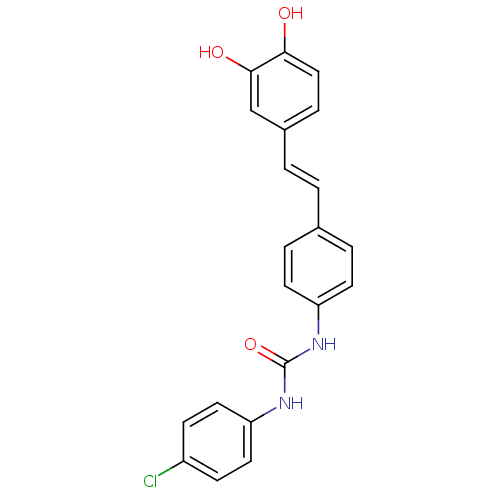
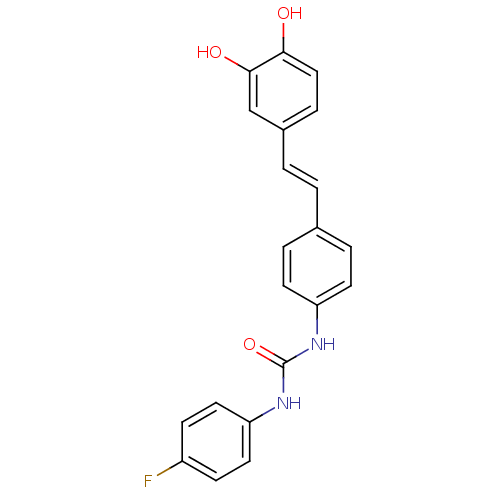
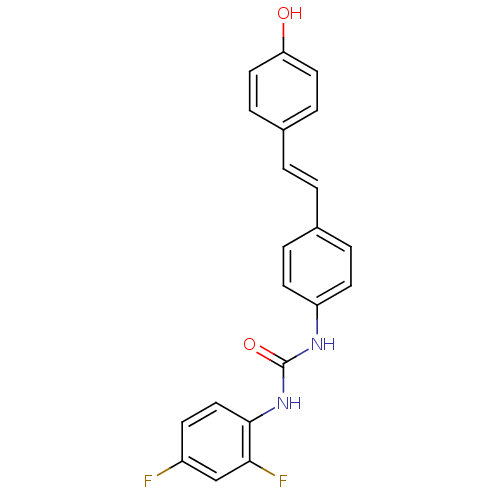
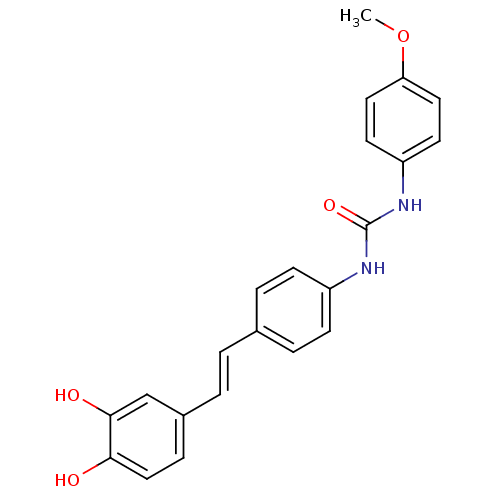
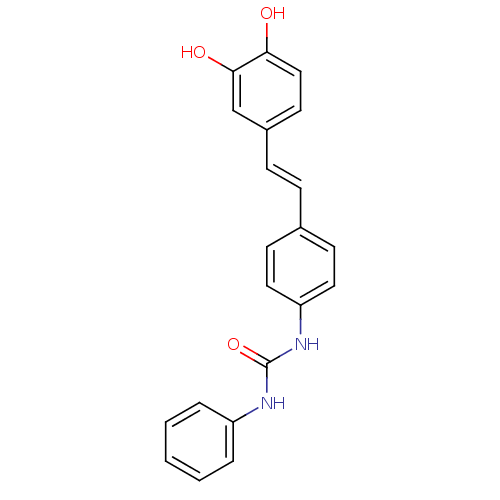
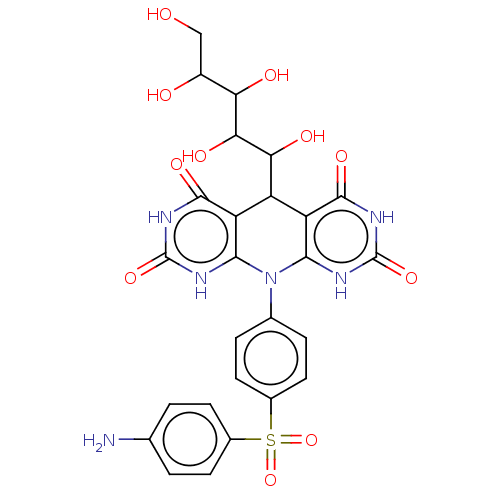
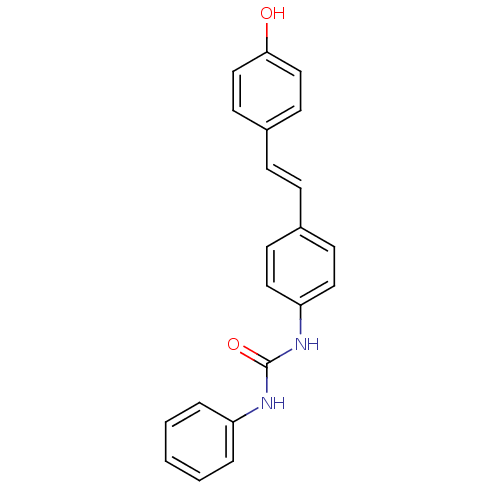
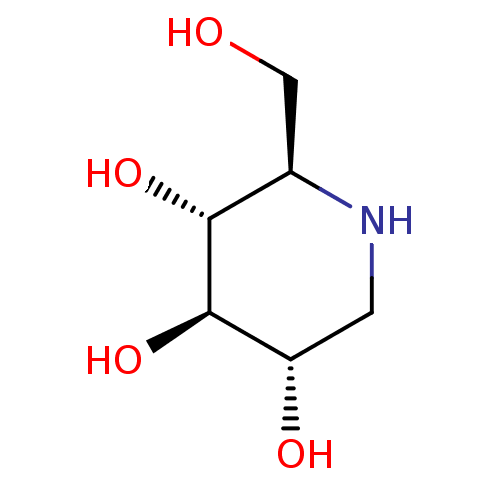
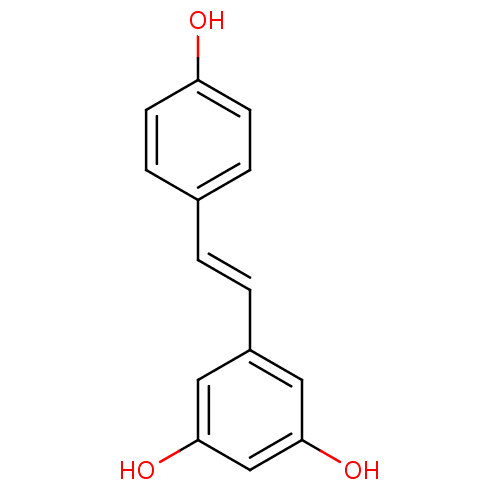
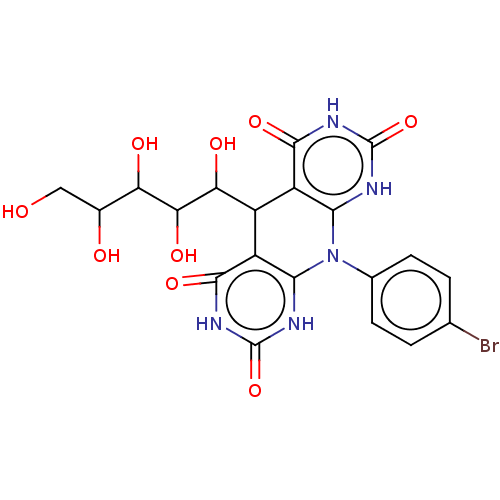
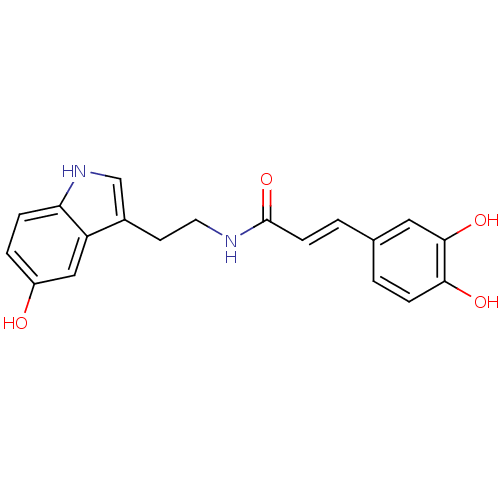
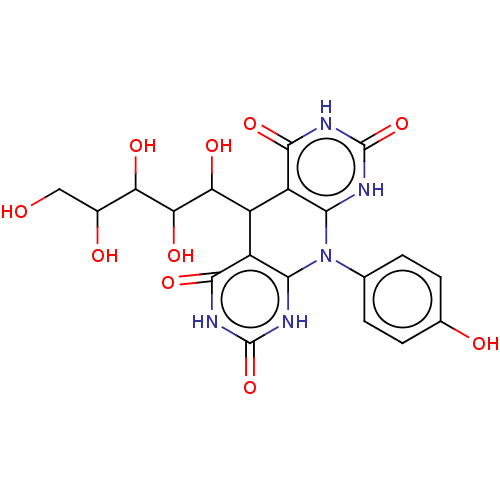
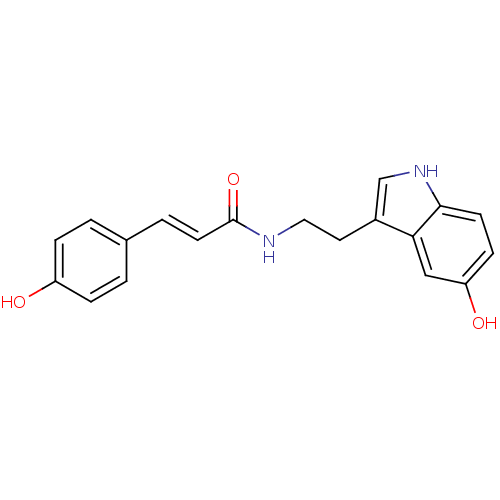
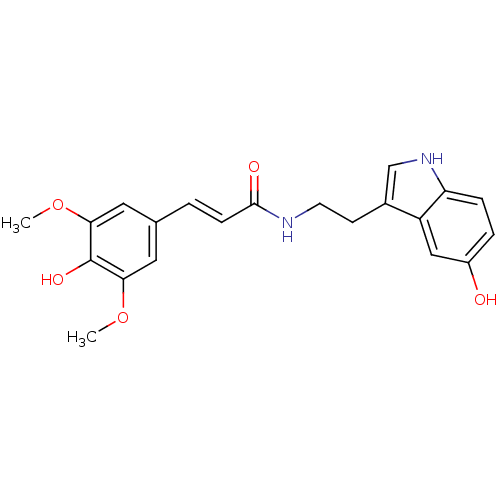
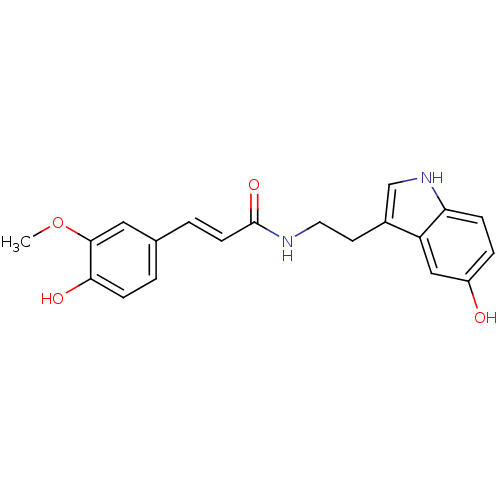
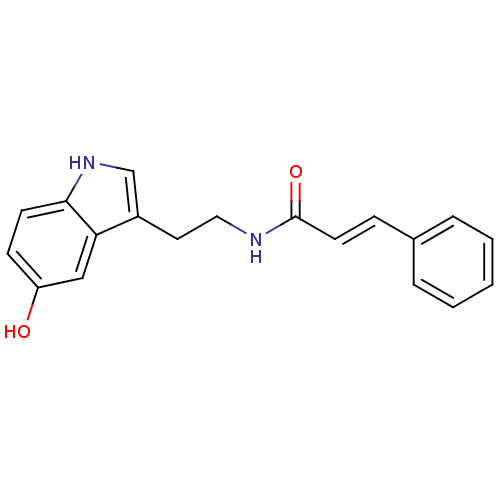
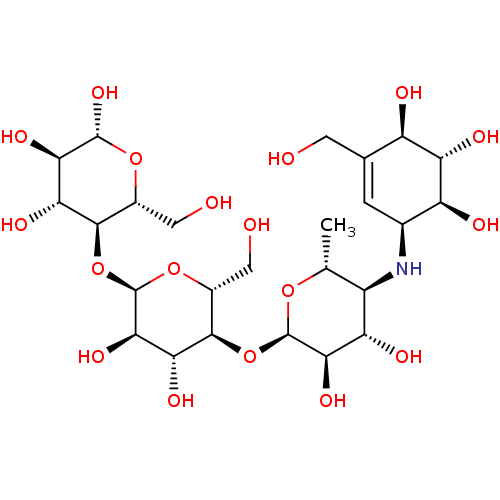
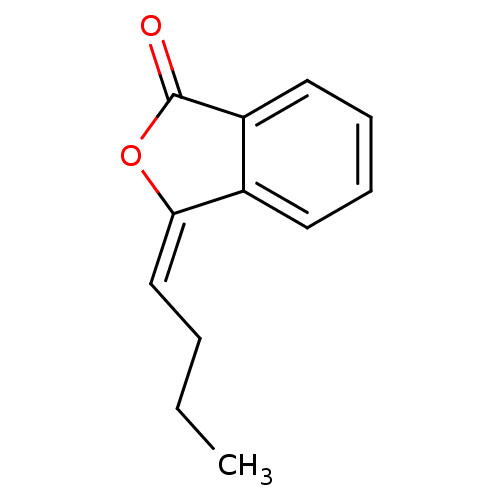
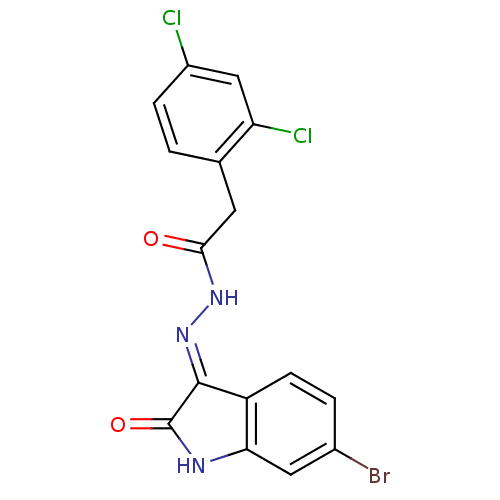
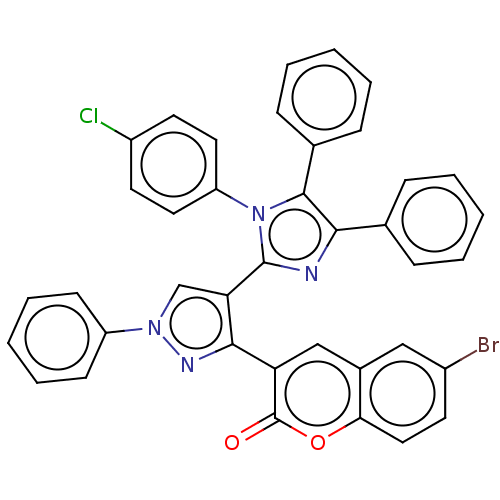
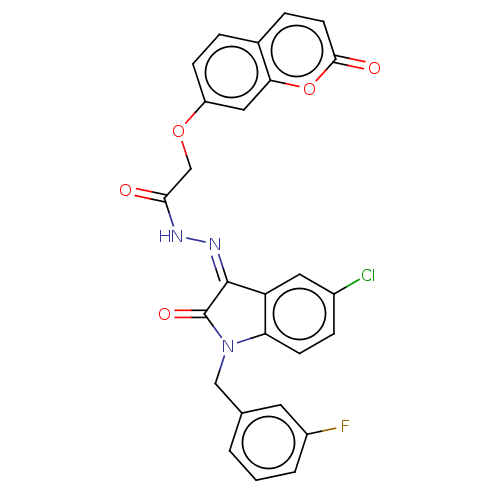
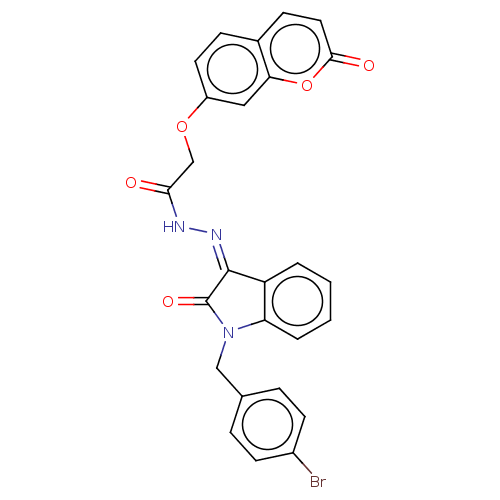
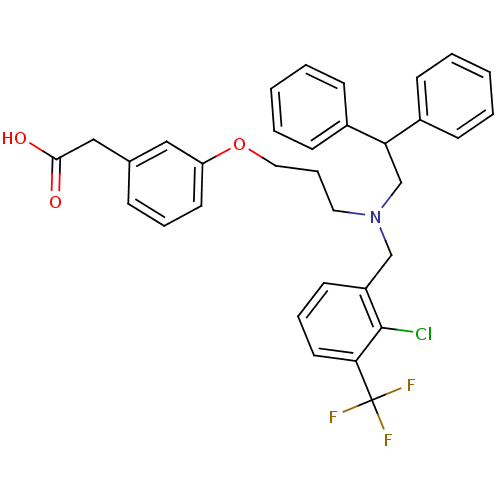
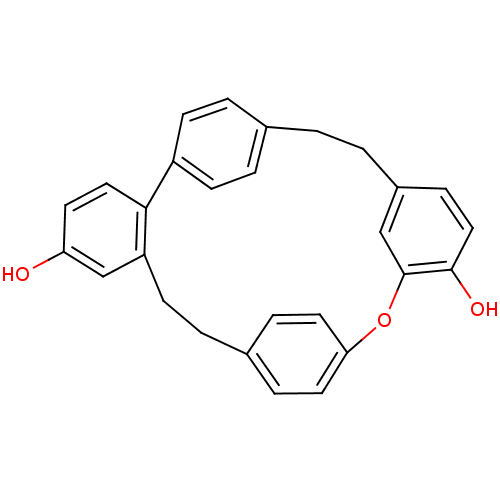
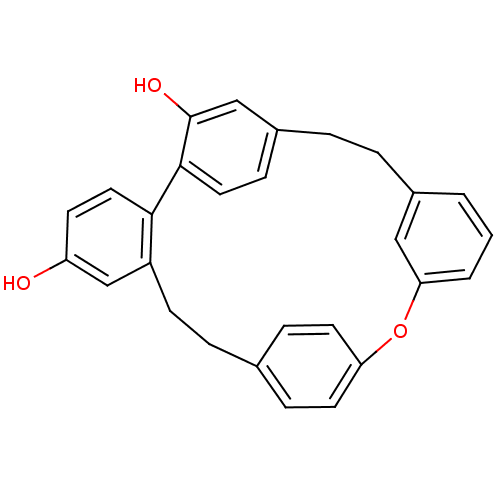
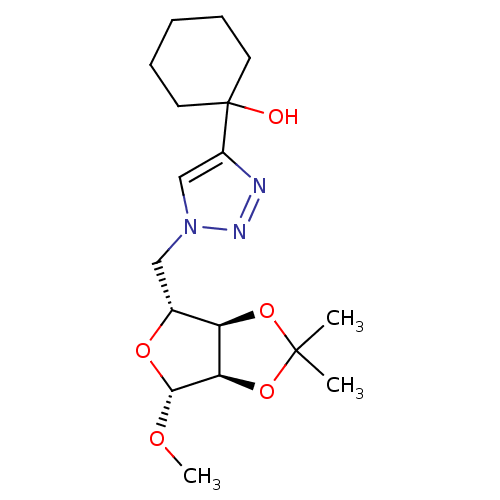
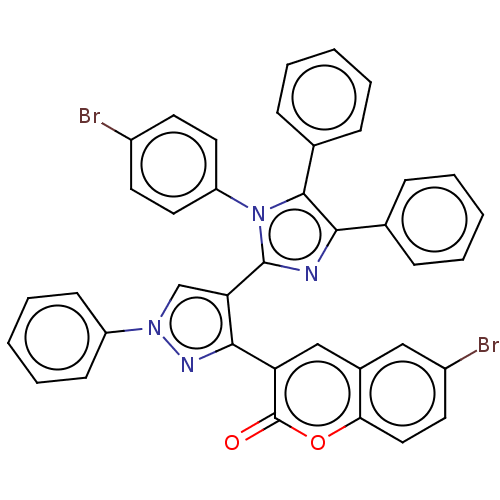
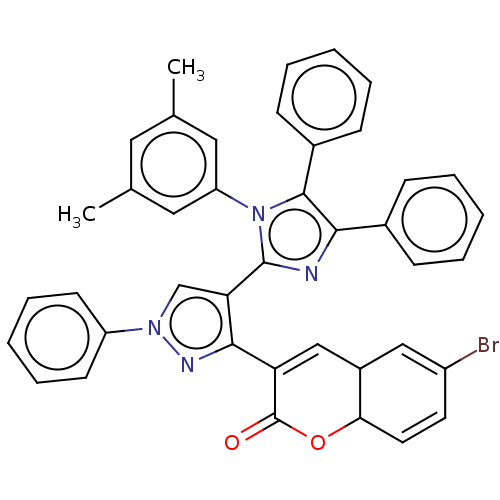
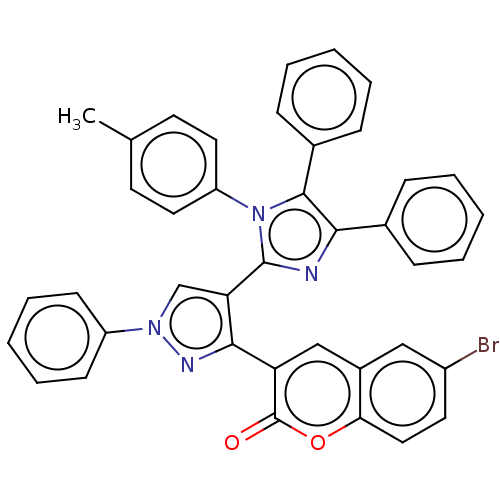
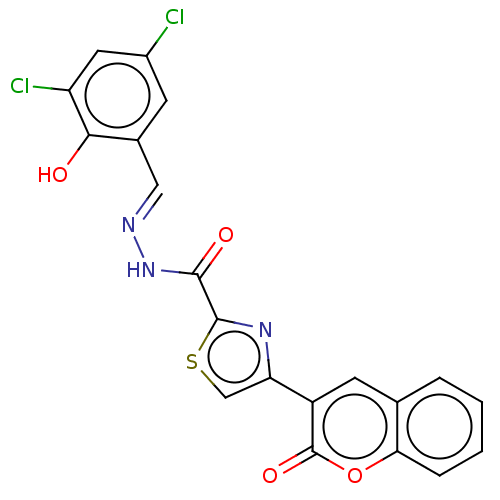
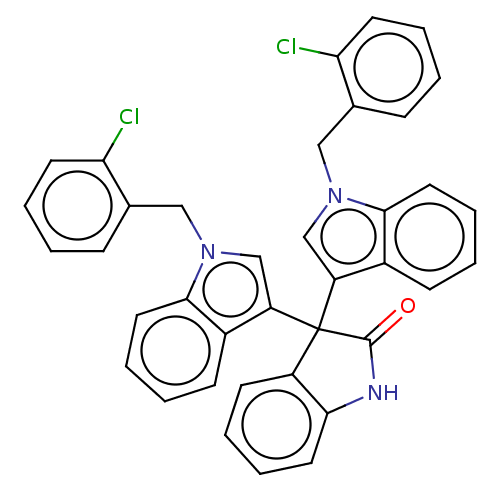
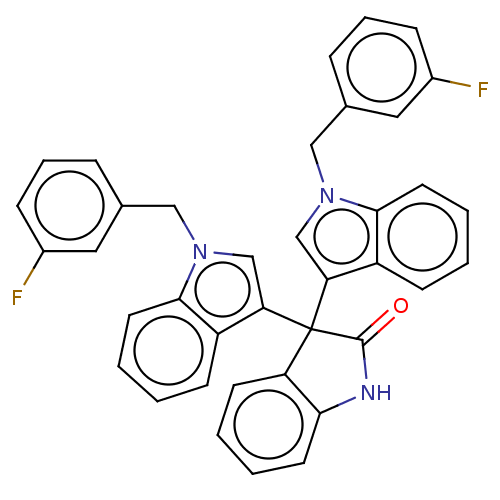
Affinity DataKi: 3.20E+3nM IC50: 8.40E+3nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
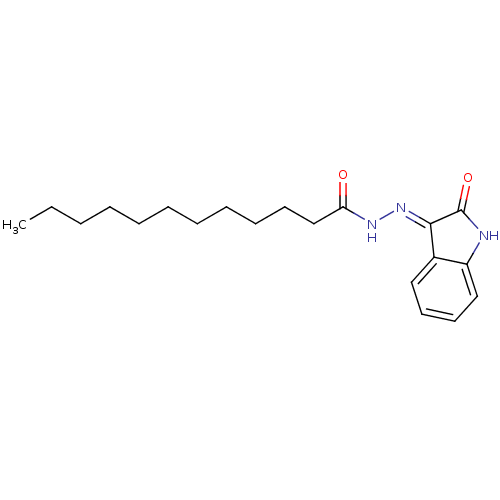
Affinity DataKi: 4.60E+3nM IC50: 1.43E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
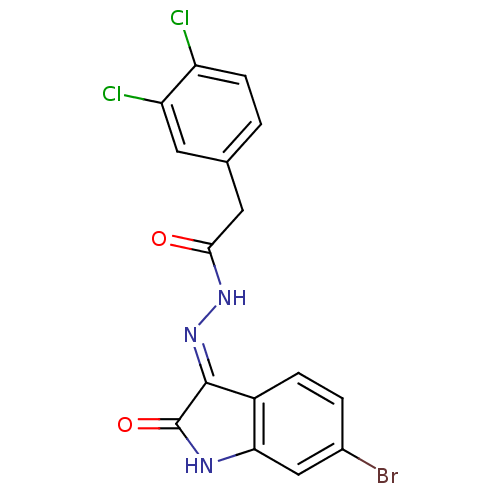
Affinity DataKi: 7.20E+3nM IC50: 1.98E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
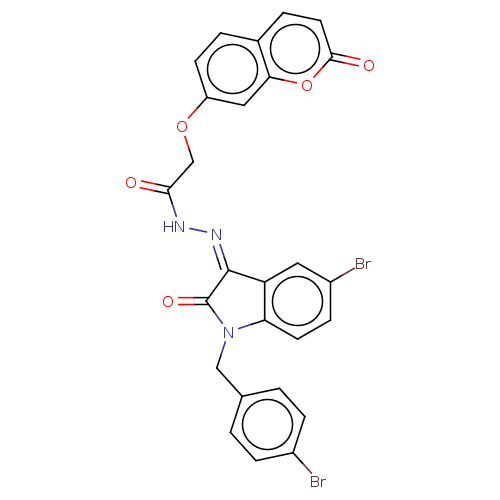
Affinity DataKi: 9.40E+3nM IC50: 1.85E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
Affinity DataKi: 1.05E+4nM IC50: 1.95E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
Affinity DataKi: 1.06E+4nM IC50: 4.21E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
Affinity DataKi: 1.21E+4nM IC50: 2.88E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
Affinity DataKi: 1.40E+4nMpH: 7.0Assay Description:In this study, the inhibition assay of yeast enzyme was performed in 100 mM phosphate buffer pH 7.0 at 25蚓 with minor changes, according to the meth...More data for this Ligand-Target Pair
Affinity DataKi: 1.70E+4nM IC50: 2.91E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
Affinity DataKi: 1.80E+4nM IC50: 3.95E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
Affinity DataKi: 2.15E+4nM IC50: 2.64E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
Affinity DataKi: 3.00E+4nMpH: 7.0Assay Description:In this study, the inhibition assay of yeast enzyme was performed in 100 mM phosphate buffer pH 7.0 at 25蚓 with minor changes, according to the meth...More data for this Ligand-Target Pair
Affinity DataKi: 3.51E+4nM IC50: 4.64E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
Affinity DataKi: 6.44E+4nMAssay Description:Uncompetitive inhibition of yeast alpha-glucosidase by Lineweaver-Burk plot analysisMore data for this Ligand-Target Pair
Affinity DataKi: 7.00E+4nMpH: 7.0Assay Description:In this study, the inhibition assay of yeast enzyme was performed in 100 mM phosphate buffer pH 7.0 at 25蚓 with minor changes, according to the meth...More data for this Ligand-Target Pair
Affinity DataKi: 8.16E+4nMAssay Description:Uncompetitive inhibition of yeast alpha-glucosidase by Lineweaver-Burk plot analysisMore data for this Ligand-Target Pair
Affinity DataKi: 1.12E+5nMAssay Description:Mixed-type inhibition of yeast alpha-glucosidase by Lineweaver-Burk plot analysisMore data for this Ligand-Target Pair
Affinity DataKi: 1.30E+5nMAssay Description:Mixed-type inhibition of yeast alpha-glucosidase by Lineweaver-Burk plot analysisMore data for this Ligand-Target Pair
Affinity DataKi: 1.30E+5nMpH: 7.0Assay Description:In this study, the inhibition assay of yeast enzyme was performed in 100 mM phosphate buffer pH 7.0 at 25蚓 with minor changes, according to the meth...More data for this Ligand-Target Pair
Affinity DataKi: 1.33E+5nMAssay Description:Competitive inhibition of yeast alpha-glucosidase by Dixon plot analysisMore data for this Ligand-Target Pair
Affinity DataKi: 1.70E+5nMpH: 7.0Assay Description:In this study, the inhibition assay of yeast enzyme was performed in 100 mM phosphate buffer pH 7.0 at 25蚓 with minor changes, according to the meth...More data for this Ligand-Target Pair
Affinity DataKi: 4.60E+5nMAssay Description:Inhibition of yeast alpha-glucosidase assessed as inhibition of p-nitrophenyl-alpha-D-glucopyranoside substrate hydrolysis after 35 mins by spectrosc...More data for this Ligand-Target Pair
Affinity DataKi: 4.86E+6nMAssay Description:Inhibition of yeast alpha-glucosidase assessed as inhibition of p-nitrophenyl-alpha-D-glucopyranoside substrate hydrolysis after 35 mins by spectrosc...More data for this Ligand-Target Pair
Affinity DataIC50: 840nMAssay Description:Inhibition of yeast maltase alpha-glucosidase assessed as p-nitrophenol release by spectrophotometricallyMore data for this Ligand-Target Pair
Affinity DataIC50: 2.00E+3nMAssay Description:Inhibition of yeast maltase alpha-glucosidase assessed as p-nitrophenol release by spectrophotometricallyMore data for this Ligand-Target Pair
Affinity DataIC50: 2.20E+3nMAssay Description:10 無 of test samples (5 mg/mL DMSO solution) were reconstituted in 100 無 of 100 mM-phosphate buffer (pH6.8) in 96-well microplate and incubated wit...More data for this Ligand-Target Pair
Affinity DataIC50: 2.53E+3nMpH: 6.8 T: 2°CAssay Description:For α-glucosidase inhibition assay, α-glucosidase enzyme (Cat No. 5003-1KU Type I; isolated from Saccharomyces cereviciae) was used. Acarbo...More data for this Ligand-Target Pair
Affinity DataIC50: 2.56E+3nMpH: 6.8Assay Description:The test compounds were dissolved in DMSO to prepare the required distributing concentration. α-Glucosidase inhibitory activity was assayed usin...More data for this Ligand-Target Pair
Affinity DataIC50: 2.85E+3nMpH: 6.8Assay Description:The test compounds were dissolved in DMSO to prepare the required distributing concentration. α-Glucosidase inhibitory activity was assayed usin...More data for this Ligand-Target Pair
Affinity DataIC50: 2.88E+3nMpH: 6.8Assay Description:The test compounds were dissolved in DMSO to prepare the required distributing concentration. α-Glucosidase inhibitory activity was assayed usin...More data for this Ligand-Target Pair
Affinity DataIC50: 3.00E+3nMAssay Description:10 無 of test samples (5 mg/mL DMSO solution) were reconstituted in 100 無 of 100 mM-phosphate buffer (pH6.8) in 96-well microplate and incubated wit...More data for this Ligand-Target Pair
Affinity DataIC50: 3.30E+3nMAssay Description:10 無 of test samples (5 mg/mL DMSO solution) were reconstituted in 100 無 of 100 mM-phosphate buffer (pH6.8) in 96-well microplate and incubated wit...More data for this Ligand-Target Pair
Affinity DataIC50: 3.80E+3nMAssay Description:Inhibition of yeast alpha-glucosidase MAL12 assessed as inhibition of p-nitrophenol release by spectrophotometryMore data for this Ligand-Target Pair
Affinity DataIC50: 3.98E+3nMpH: 6.8Assay Description:The test compounds were dissolved in DMSO to prepare the required distributing concentration. α-Glucosidase inhibitory activity was assayed usin...More data for this Ligand-Target Pair
Affinity DataIC50: 4.07E+3nMpH: 6.8Assay Description:The test compounds were dissolved in DMSO to prepare the required distributing concentration. α-Glucosidase inhibitory activity was assayed usin...More data for this Ligand-Target Pair
Affinity DataIC50: 4.48E+3nMpH: 6.8Assay Description:The test compounds were dissolved in DMSO to prepare the required distributing concentration. α-Glucosidase inhibitory activity was assayed usin...More data for this Ligand-Target Pair
Affinity DataIC50: 4.80E+3nMpH: 7.0 T: 2°CAssay Description:The alpha-glucosidase inhibitory activity of test compounds was determined in a 96-well plate format. The reaction mixture containing enzyme and chro...More data for this Ligand-Target Pair
Affinity DataIC50: 4.90E+3nMpH: 7.0 T: 2°CAssay Description:The alpha-glucosidase inhibitory activity of test compounds was determined in a 96-well plate format. The reaction mixture containing enzyme and chro...More data for this Ligand-Target Pair
Affinity DataIC50: 5.10E+3nMpH: 7.0 T: 2°CAssay Description:The alpha-glucosidase inhibitory activity of test compounds was determined in a 96-well plate format. The reaction mixture containing enzyme and chro...More data for this Ligand-Target Pair
Affinity DataIC50: 5.20E+3nMAssay Description:Inhibition of yeast alpha-glucosidase MAL12 assessed as inhibition of p-nitrophenol release by spectrophotometryMore data for this Ligand-Target Pair
Affinity DataIC50: 5.21E+3nMpH: 6.8 T: 2°CAssay Description:For α-glucosidase inhibition assay, α-glucosidase enzyme (Cat No. 5003-1KU Type I; isolated from Saccharomyces cereviciae) was used. Acarbo...More data for this Ligand-Target Pair
Affinity DataIC50: 5.64E+3nMpH: 6.8 T: 2°CAssay Description:For α-glucosidase inhibition assay, α-glucosidase enzyme (Cat No. 5003-1KU Type I; isolated from Saccharomyces cereviciae) was used. Acarbo...More data for this Ligand-Target Pair
Affinity DataIC50: 5.70E+3nMAssay Description:Inhibition of yeast alpha-glucosidase MAL12 assessed as inhibition of p-nitrophenol release by spectrophotometryMore data for this Ligand-Target Pair
Affinity DataIC50: 5.83E+3nMpH: 6.8 T: 2°CAssay Description:For α-glucosidase inhibition assay, α-glucosidase enzyme (Cat No. 5003-1KU Type I; isolated from Saccharomyces cereviciae) was used. Acarbo...More data for this Ligand-Target Pair
Affinity DataIC50: 5.98E+3nMpH: 6.8 T: 2°CAssay Description:The α-glucosidase inhibition assay had been carried out using baker's yeast α-glucosidase (EC 3.2.1.20) and p-nitrophenyl α-d-gluco...More data for this Ligand-Target Pair
Affinity DataIC50: 6.24E+3nMpH: 6.8Assay Description:The test compounds were dissolved in DMSO to prepare the required distributing concentration. α-Glucosidase inhibitory activity was assayed by u...More data for this Ligand-Target Pair
Affinity DataIC50: 7.31E+3nMpH: 6.8Assay Description:The test compounds were dissolved in DMSO to prepare the required distributing concentration. α-Glucosidase inhibitory activity was assayed usin...More data for this Ligand-Target Pair
Affinity DataIC50: 7.31E+3nMpH: 6.8 T: 2°CAssay Description:The α-glucosidase inhibition assay had been carried out using baker's yeast α-glucosidase (EC 3.2.1.20) and p-nitrophenyl α-d-gluco...More data for this Ligand-Target Pair
Affinity DataIC50: 7.37E+3nMpH: 6.8Assay Description:The test compounds were dissolved in DMSO to prepare the required distributing concentration. α-Glucosidase inhibitory activity was assayed usin...More data for this Ligand-Target Pair
Affinity DataIC50: 7.92E+3nMpH: 6.8 T: 2°CAssay Description:The α-glucosidase inhibition assay had been carried out using baker's yeast α-glucosidase (EC 3.2.1.20) and p-nitrophenyl α-d-gluco...More data for this Ligand-Target Pair