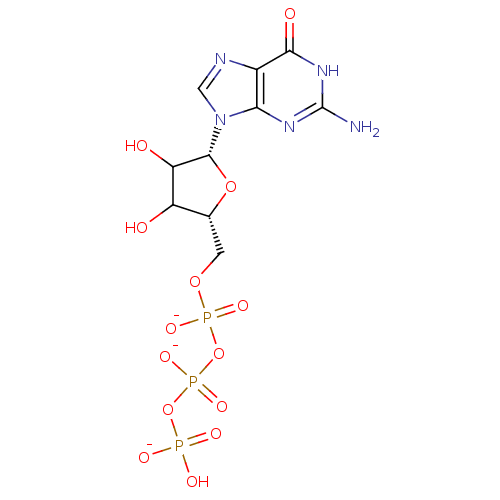
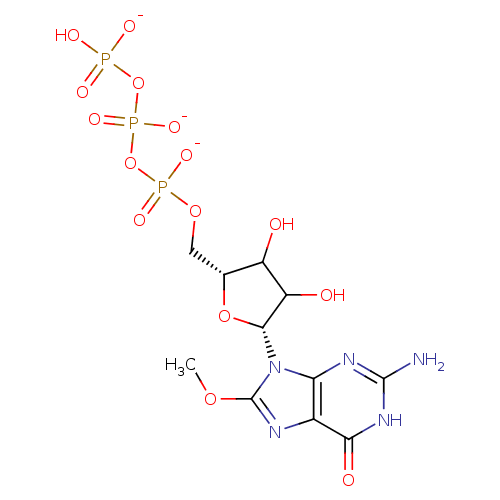
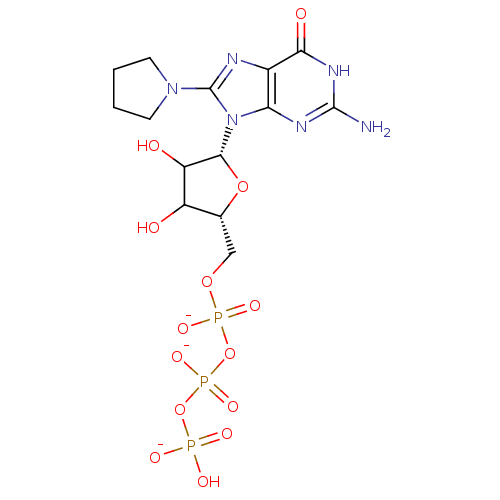
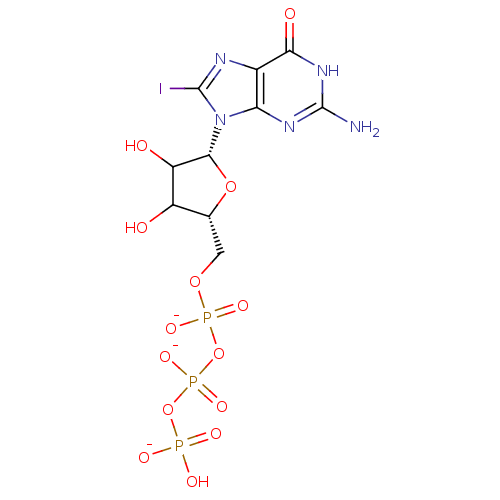
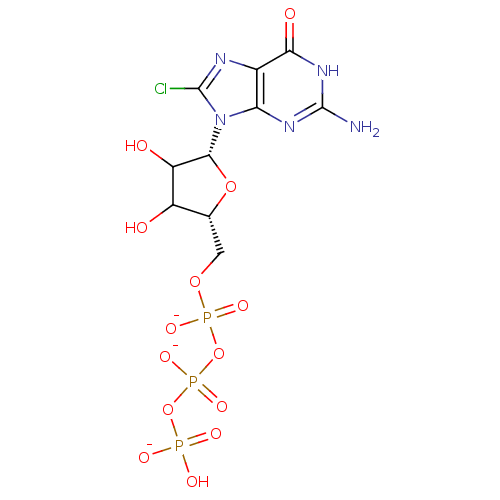
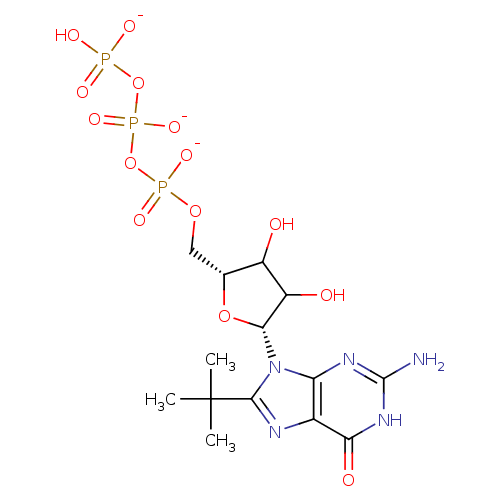
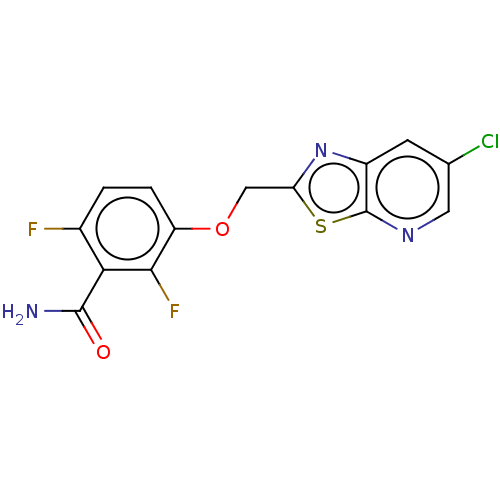
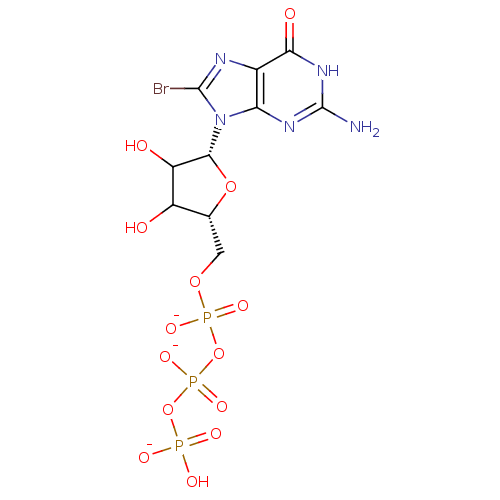
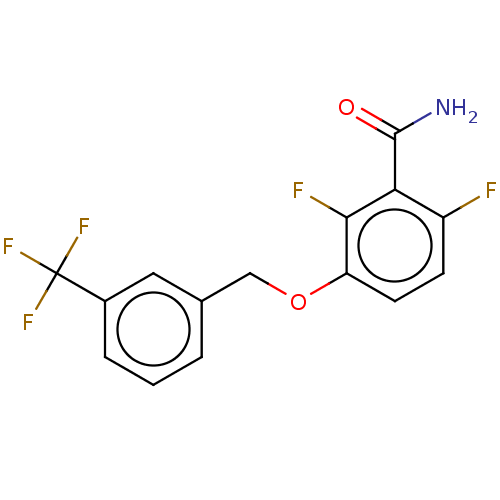
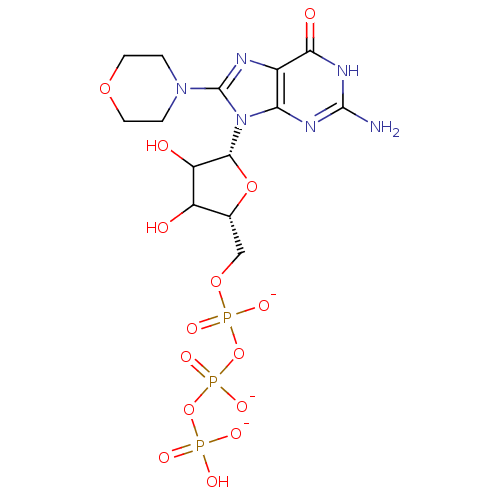
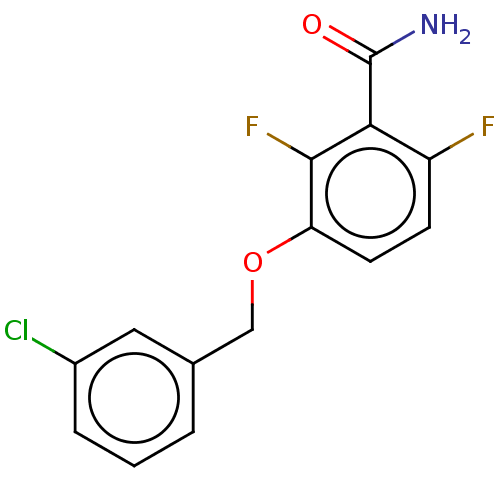
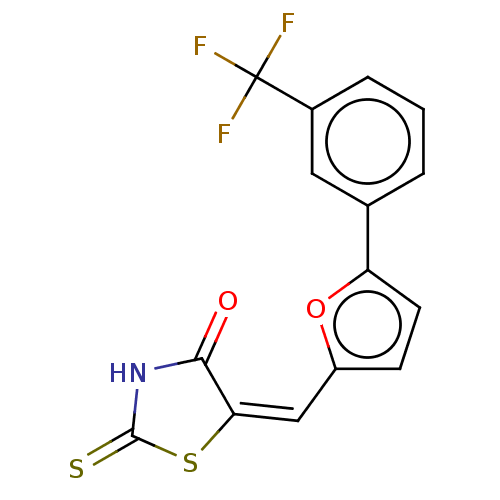
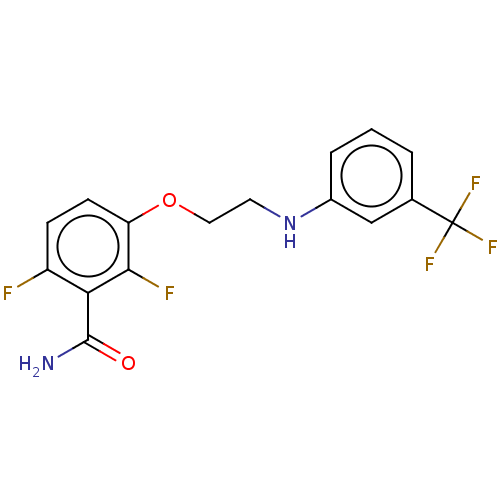
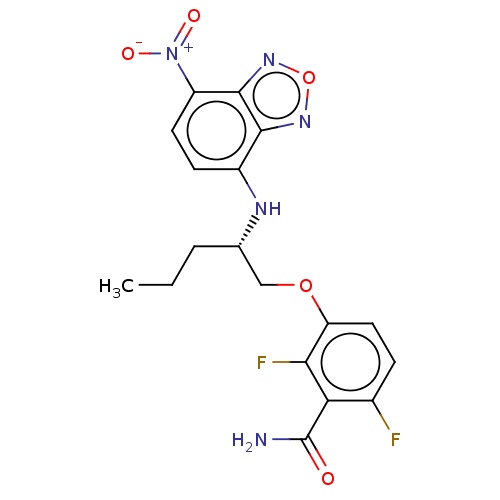
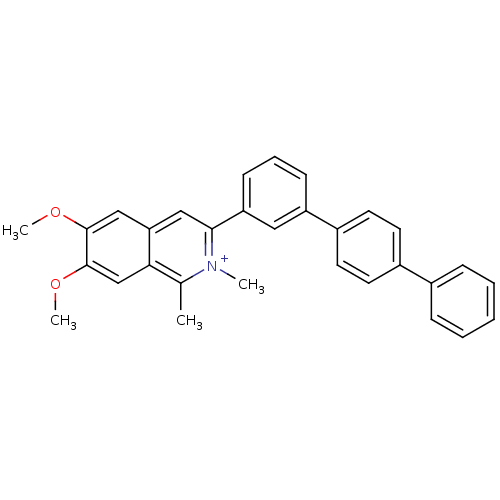
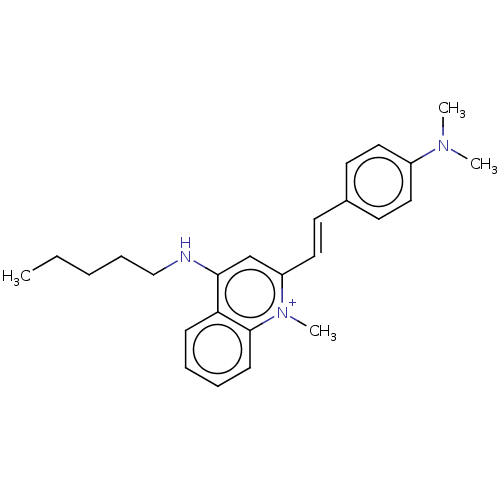
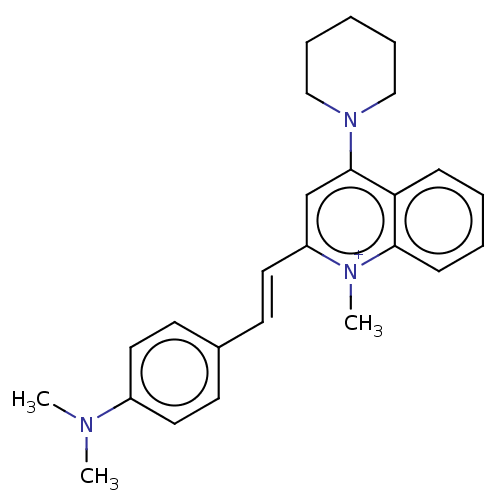
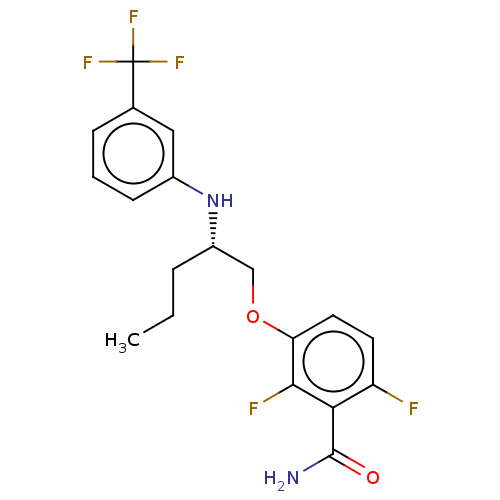
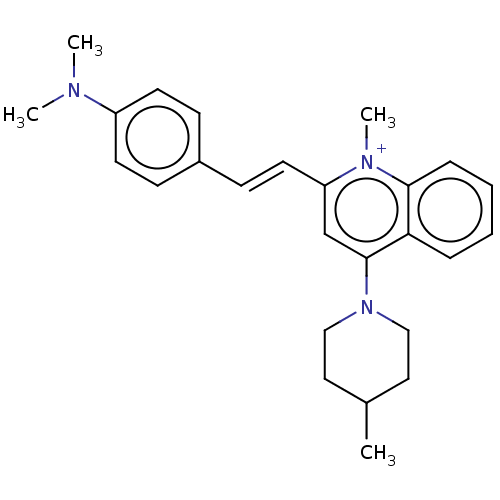
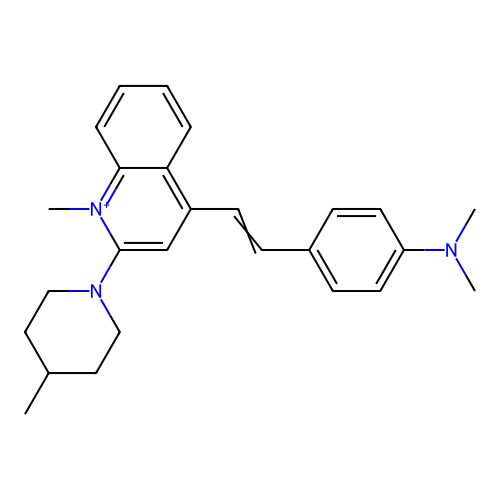
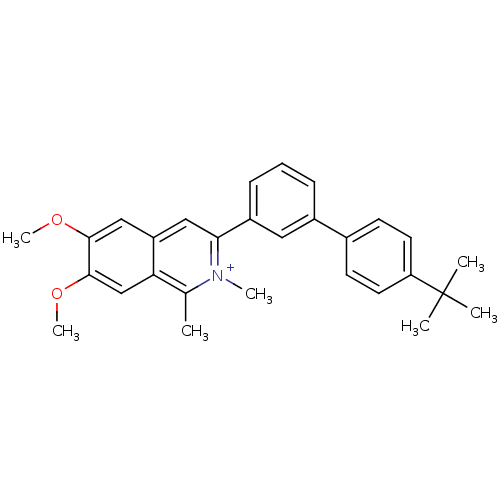
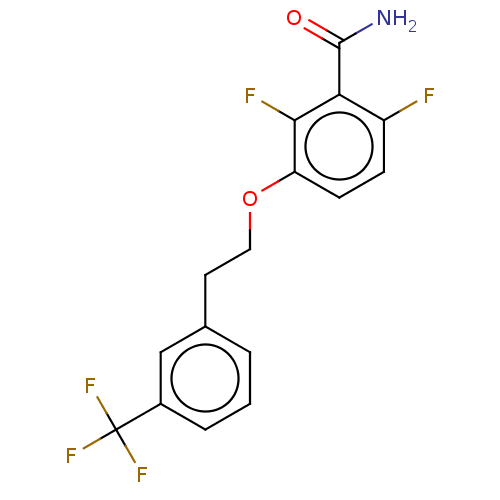
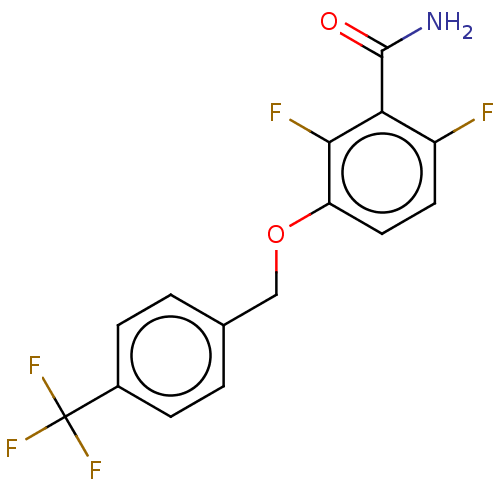
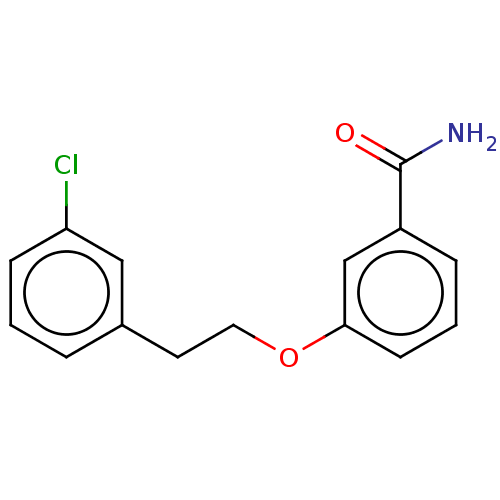
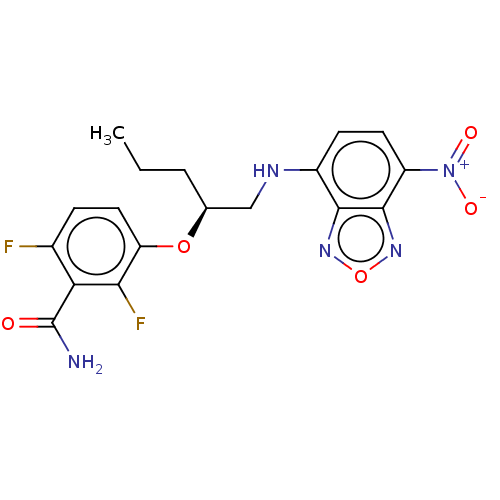
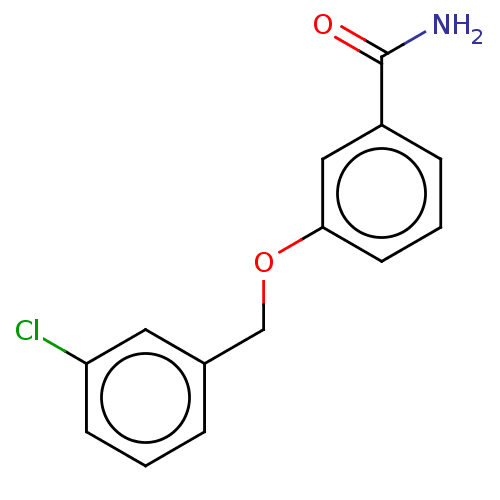
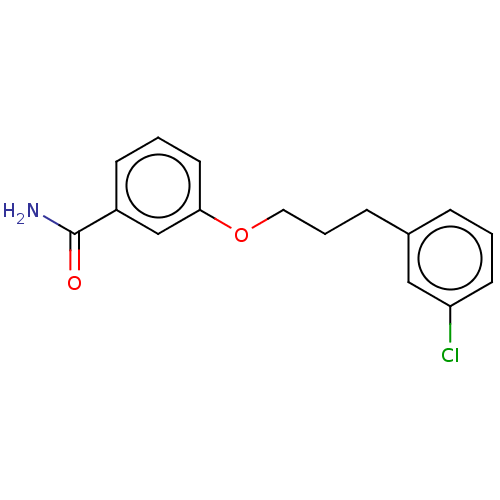
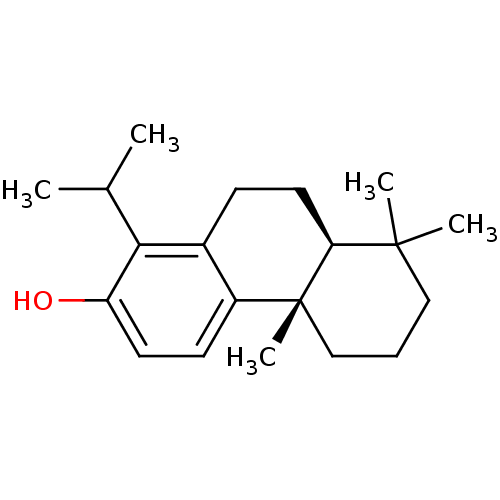
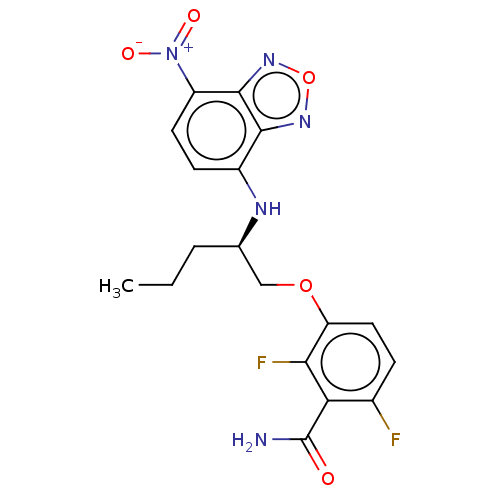
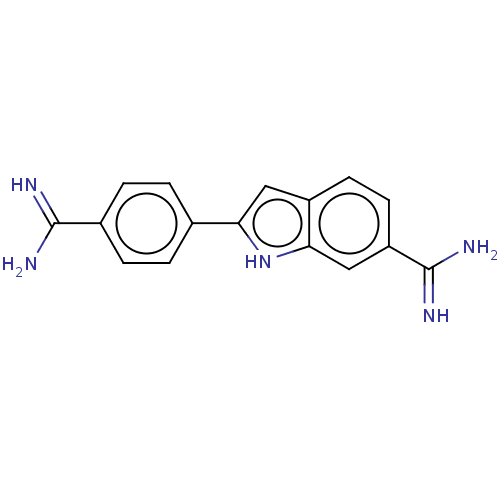
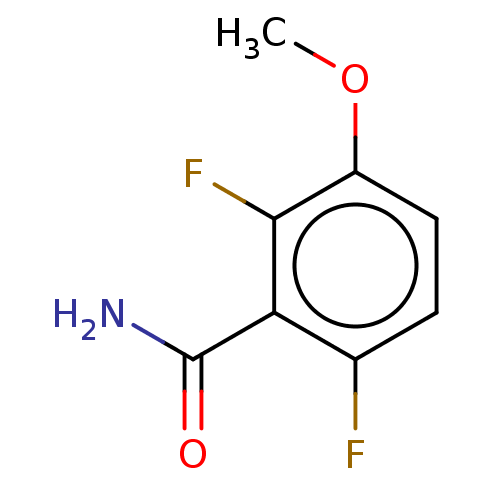
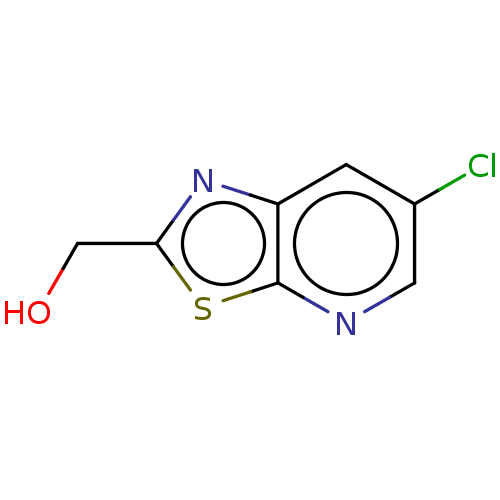
Affinity DataKd: 3.03nMpH: 7.5Assay Description:The binding affinities to FtsZ were determined by a fluorescent competition assay with 2'/3'-O-(N-methyl-anthraniloyl)-guanosine-5'-triph...More data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+4nM Kd: 3.33nMpH: 7.5Assay Description:The binding affinities to FtsZ were determined by a fluorescent competition assay with 2'/3'-O-(N-methyl-anthraniloyl)-guanosine-5'-triph...More data for this Ligand-Target Pair
Affinity DataIC50: 1.50E+4nM Kd: 15.6nMpH: 7.5Assay Description:The binding affinities to FtsZ were determined by a fluorescent competition assay with 2'/3'-O-(N-methyl-anthraniloyl)-guanosine-5'-triph...More data for this Ligand-Target Pair
Affinity DataKd: 23nMAssay Description:Displacement of bis-ANS from Escherichia coli FtsZMore data for this Ligand-Target Pair
Affinity DataIC50: 4.40E+4nM Kd: 71nMpH: 7.5Assay Description:The binding affinities to FtsZ were determined by a fluorescent competition assay with 2'/3'-O-(N-methyl-anthraniloyl)-guanosine-5'-triph...More data for this Ligand-Target Pair
Affinity DataIC50: 3.50E+4nM Kd: 77nMpH: 7.5Assay Description:The binding affinities to FtsZ were determined by a fluorescent competition assay with 2'/3'-O-(N-methyl-anthraniloyl)-guanosine-5'-triph...More data for this Ligand-Target Pair
Affinity DataIC50: 4.10E+4nM Kd: 91nMpH: 7.5Assay Description:The binding affinities to FtsZ were determined by a fluorescent competition assay with 2'/3'-O-(N-methyl-anthraniloyl)-guanosine-5'-triph...More data for this Ligand-Target Pair
Affinity DataKd: <100nMAssay Description:Displacement of free fluorescent probe from inter domain cleft of stabilized Bacillus subtilis FtsZ assessed as dissociation constant in presence of ...More data for this Ligand-Target Pair
Affinity DataIC50: 3.70E+4nM Kd: 126nMpH: 7.5Assay Description:The binding affinities to FtsZ were determined by a fluorescent competition assay with 2'/3'-O-(N-methyl-anthraniloyl)-guanosine-5'-triph...More data for this Ligand-Target Pair
Affinity DataKd: 200nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Staphylococcus aureus)
The Hong Kong Polytechnic University
Curated by ChEMBL
The Hong Kong Polytechnic University
Curated by ChEMBL
Affinity DataKd: 600nMAssay Description:Binding affinity to Staphylococcus aureus FtsZ assessed as dissociation constant by isothermal titration calorimetryMore data for this Ligand-Target Pair
Affinity DataKd: 700nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 1.39E+5nM Kd: 714nMpH: 7.5Assay Description:The binding affinities to FtsZ were determined by a fluorescent competition assay with 2'/3'-O-(N-methyl-anthraniloyl)-guanosine-5'-triph...More data for this Ligand-Target Pair
Affinity DataKd: 1.10E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
Affinity DataKd: 1.30E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
Affinity DataKd: 1.50E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
Affinity DataKd: 1.50E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZMore data for this Ligand-Target Pair
Affinity DataKd: 1.90E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
Affinity DataKd: 1.90E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Staphylococcus aureus)
The Hong Kong Polytechnic University
Curated by ChEMBL
The Hong Kong Polytechnic University
Curated by ChEMBL
Affinity DataKd: 2.00E+3nMAssay Description:Binding affinity to Staphylococcus aureus FtsZ expressed in Escherichia coli cells assessed as dissociation constant by fluorescence spectroscopyMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Staphylococcus aureus)
The Hong Kong Polytechnic University
Curated by ChEMBL
The Hong Kong Polytechnic University
Curated by ChEMBL
Affinity DataKd: 2.38E+3nMAssay Description:Binding affinity to Staphylococcus aureus FtsZ assessed as dissociation constant by isothermal titration calorimetryMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Staphylococcus aureus)
The Hong Kong Polytechnic University
Curated by ChEMBL
The Hong Kong Polytechnic University
Curated by ChEMBL
Affinity DataKd: 2.49E+3nMAssay Description:Binding affinity to Staphylococcus aureus FtsZ assessed as dissociation constant by isothermal titration calorimetryMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Staphylococcus aureus)
The Hong Kong Polytechnic University
Curated by ChEMBL
The Hong Kong Polytechnic University
Curated by ChEMBL
Affinity DataKd: 3.50E+3nMAssay Description:Binding affinity to Staphylococcus aureus FtsZ expressed in Escherichia coli BL21 (DE3) at 25 degC by fluorescence anisotropy assayMore data for this Ligand-Target Pair
Affinity DataKd: 3.80E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
Affinity DataKd: 3.80E+3nMAssay Description:Binding affinity to Escherichia coli FtsZ at 25 degC by fluorescence anisotropy assayMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Staphylococcus aureus)
The Hong Kong Polytechnic University
Curated by ChEMBL
The Hong Kong Polytechnic University
Curated by ChEMBL
Affinity DataKd: 3.84E+3nMAssay Description:Binding affinity to Staphylococcus aureus FtsZ assessed as dissociation constant by isothermal titration calorimetryMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Staphylococcus aureus)
The Hong Kong Polytechnic University
Curated by ChEMBL
The Hong Kong Polytechnic University
Curated by ChEMBL
Affinity DataKd: 3.86E+3nMAssay Description:Binding affinity to Staphylococcus aureus FtsZ assessed as dissociation constant by isothermal titration calorimetryMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Staphylococcus aureus)
The Hong Kong Polytechnic University
Curated by ChEMBL
The Hong Kong Polytechnic University
Curated by ChEMBL
Affinity DataKd: 5.40E+3nMAssay Description:Binding affinity to Staphylococcus aureus FtsZ expressed in Escherichia coli cells assessed as dissociation constant by fluorescence spectroscopyMore data for this Ligand-Target Pair
Affinity DataKd: 5.90E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
Affinity DataKd: 6.70E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
Affinity DataKd: 7.10E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
Affinity DataKd: 8.00E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
Affinity DataKd: 9.30E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
Affinity DataKd: 9.50E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
Affinity DataKd: 1.10E+4nMAssay Description:Binding affinity to Mycobacterium tuberculosis FtsZMore data for this Ligand-Target Pair
Affinity DataKd: 1.50E+4nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
Affinity DataKd: 1.66E+4nMAssay Description:Binding affinity to Escherichia coli FtsZ by fluorescence anisotrophyMore data for this Ligand-Target Pair
Affinity DataKd: 7.10E+4nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
Affinity DataKd: 1.45E+5nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
Affinity DataKd: 6.70E+5nMAssay Description:Displacement of free fluorescent probe from inter domain cleft of stabilized Bacillus subtilis FtsZ assessed as dissociation constant in presence of ...More data for this Ligand-Target Pair
Affinity DataKd: 1.00E+6nMAssay Description:Displacement of free fluorescent probe from inter domain cleft of stabilized Bacillus subtilis FtsZ assessed as dissociation constant in presence of ...More data for this Ligand-Target Pair
TargetCell division protein FtsZ(Staphylococcus aureus)
The Hong Kong Polytechnic University
Curated by ChEMBL
The Hong Kong Polytechnic University
Curated by ChEMBL
Affinity DataIC50: 6.70E+4nMAssay Description:Inhibition of recombinant Staphylococcus aureus FtsZ GTPase activity expressed in Escherichia coli BL21 (DE3) assessed as production of inorganic pho...More data for this Ligand-Target Pair
TargetCell division protein FtsZ(Staphylococcus aureus)
The Hong Kong Polytechnic University
Curated by ChEMBL
The Hong Kong Polytechnic University
Curated by ChEMBL
Affinity DataIC50: 3.80E+4nMAssay Description:Inhibition of recombinant Staphylococcus aureus FtsZ GTPase activity expressed in Escherichia coli BL21 (DE3) assessed as production of inorganic pho...More data for this Ligand-Target Pair
Affinity DataIC50: 7.00E+4nMAssay Description:Inhibition of Bacillus subtilis FtsZ GTPase activity assessed as production of inorganic phosphate measured up to 60 mins by malachite green-phosphom...More data for this Ligand-Target Pair
Affinity DataIC50: 1.53E+4nMpH: 7.5Assay Description:The binding affinities to FtsZ were determined by a fluorescent coupled assay for phosphate relase. In this GTPase studies, phosphate liberated durin...More data for this Ligand-Target Pair
Affinity DataIC50: 2.14E+4nMpH: 7.5Assay Description:The binding affinities to FtsZ were determined by a fluorescent coupled assay for phosphate relase. In this GTPase studies, phosphate liberated durin...More data for this Ligand-Target Pair
Affinity DataIC50: 4.40E+4nMpH: 7.5Assay Description:The binding affinities to FtsZ were determined by a fluorescent coupled assay for phosphate relase. In this GTPase studies, phosphate liberated durin...More data for this Ligand-Target Pair
Affinity DataIC50: 4.40E+4nMpH: 7.5Assay Description:The binding affinities to FtsZ were determined by a fluorescent competition assay with 2'/3'-O-(N-methyl-anthraniloyl)-guanosine-5'-triph...More data for this Ligand-Target Pair
Affinity DataIC50: 5.20E+4nMpH: 7.5Assay Description:The binding affinities to FtsZ were determined by a fluorescent coupled assay for phosphate relase. In this GTPase studies, phosphate liberated durin...More data for this Ligand-Target Pair
Affinity DataIC50: 6.02E+4nMpH: 7.5Assay Description:The binding affinities to FtsZ were determined by a fluorescent coupled assay for phosphate relase. In this GTPase studies, phosphate liberated durin...More data for this Ligand-Target Pair