Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
ATP-dependent molecular chaperone HSP82
Ligand
BDBM64876
Substrate
n/a
Meas. Tech.
Fluorescence Cell-Based Secondary Assay to Identify Inhibitors of Resistant C. albicans Growth in the Presence of Fluconazole
EC50
7573±1439 nM
Citation
 PubChem, PC Fluorescence Cell-Based Secondary Assay to Identify Inhibitors of Resistant C. albicans Growth in the Presence of Fluconazole PubChem Bioassay (2010)[AID]
PubChem, PC Fluorescence Cell-Based Secondary Assay to Identify Inhibitors of Resistant C. albicans Growth in the Presence of Fluconazole PubChem Bioassay (2010)[AID] More Info.:
Target
Name:
ATP-dependent molecular chaperone HSP82
Synonyms:
heat shock protein 90
Type:
Enzyme Catalytic Domain
Mol. Mass.:
80784.12
Organism:
Candida albicans
Description:
gi_994798
Residue:
707
Sequence:
MADAKVETHEFTAEISQLMSLIINTVYSNKEIFLRELISNASDALDKIRYQALSDPSQLESEPELFIRIIPQKDQKVLEIRDSGIGMTKADLVNNLGTIAKSGTKSFMEALSAGADVSMIGQFGVGFYSLFLVADHVQVISKHNDDEQYVWESNAGGKFTVTLDETNERLGRGTMLRLFLKEDQLEYLEEKRIKEVVKKHSEFVAYPIQLVVTKEVEKEVPETEEEDKAAEEDDKKPKLEEVKDEEDEKKEKKTKTVKEEVTETEELNKTKPLWTRNPSDITQDEYNAFYKSISNDWEDPLAVKHFSVEGQLEFRAILFVPKRAPFDAFESKKKKNNIKLYVRRVFITDDAEELIPEWLSFIKGVVDSEDLPLNLSREMLQQNKILKVIRKNIVKKMIETFNEISEDQEQFNQFYTAFSKNIKLGIHEDAQNRQSLAKLLRFYSTKSSEEMTSLSDYVTRMPEHQKNIYYITGESIKAVEKSPFLDALKAKNFEVLFMVDPIDEYAMTQLKEFEDKKLVDITKDFELEESDEEKAAREKEIKEYEPLTKALKDILGDQVEKVVVSYKLVDAPAAIRTGQFGWSANMERIMKAQALRDTTMSSYMSSKKTFEISPSSPIIKELKKKVETDGAEDKTVKDLTTLLFDTALLTSGFTLDEPSNFAHRINRLIALGLNIDDDSEETAVEPEATTTASTDEPAGESAMEEVD
Inhibitor
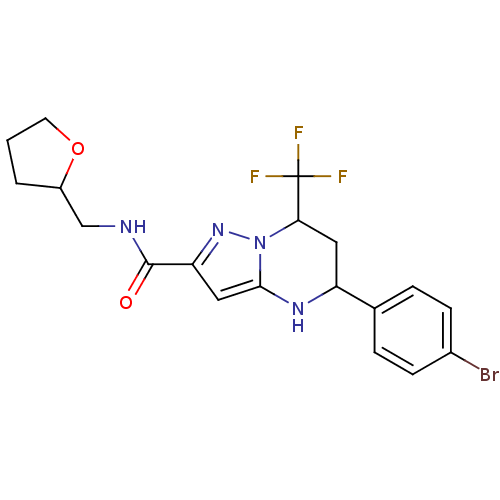
Name:
BDBM64876
Synonyms:
5-(4-Bromo-phenyl)-7-trifluoromethyl-4,5,6,7-tetrahydro-pyrazolo[1,5-a]pyrimidine-2-carboxylic acid (tetrahydro-furan-2-ylmethyl)-amide | 5-(4-bromophenyl)-N-(2-oxolanylmethyl)-7-(trifluoromethyl)-1,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-2-carboxamide | 5-(4-bromophenyl)-N-(oxolan-2-ylmethyl)-7-(trifluoromethyl)-1,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-2-carboxamide | 5-(4-bromophenyl)-N-(tetrahydrofurfuryl)-7-(trifluoromethyl)-1,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-2-carboxamide | MLS000070394 | SMR000010783 | cid_5764472
Type:
Small organic molecule
Emp. Form.:
C19H20BrF3N4O2
Mol. Mass.:
473.287
SMILES:
FC(F)(F)C1CC(Nc2cc(nn12)C(=O)NCC1CCCO1)c1ccc(Br)cc1