Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Serine protease 1/Trypsin-2
Ligand
BDBM50102780
Substrate
n/a
Meas. Tech.
ChEMBL_225794 (CHEMBL845144)
Ki
130±n/a nM
Citation
 Mackman, RL; Katz, BA; Breitenbucher, JG; Hui, HC; Verner, E; Luong, C; Liu, L; Sprengeler, PA Exploiting subsite S1 of trypsin-like serine proteases for selectivity: potent and selective inhibitors of urokinase-type plasminogen activator. J Med Chem 44:3856-71 (2001) [PubMed] Article
Mackman, RL; Katz, BA; Breitenbucher, JG; Hui, HC; Verner, E; Luong, C; Liu, L; Sprengeler, PA Exploiting subsite S1 of trypsin-like serine proteases for selectivity: potent and selective inhibitors of urokinase-type plasminogen activator. J Med Chem 44:3856-71 (2001) [PubMed] Article More Info.:
Target
Name:
Serine protease 1/Trypsin-2
Synonyms:
Trypsin II
Type:
n/a
Mol. Mass.:
n/a
Description:
ASSAY_ID of EBI is 212335
Components:
This complex has 2 components.
Component 1
Name:
Serine protease 1
Synonyms:
Alpha-trypsin chain 1 | Alpha-trypsin chain 2 | Beta-trypsin | Cationic trypsinogen | PRSS1 | Serine protease 1 | TRP1 | TRY1 | TRY1_HUMAN | TRYP1 | Thrombin & trypsin | Trypsin | Trypsin I | Trypsin-1
Type:
Enzyme
Mol. Mass.:
26557.80
Organism:
Homo sapiens (Human)
Description:
P07477
Residue:
247
Sequence:
MNPLLILTFVAAALAAPFDDDDKIVGGYNCEENSVPYQVSLNSGYHFCGGSLINEQWVVSAGHCYKSRIQVRLGEHNIEVLEGNEQFINAAKIIRHPQYDRKTLNNDIMLIKLSSRAVINARVSTISLPTAPPATGTKCLISGWGNTASSGADYPDELQCLDAPVLSQAKCEASYPGKITSNMFCVGFLEGGKDSCQGDSGGPVVCNGQLQGVVSWGDGCAQKNKPGVYTKVYNYVKWIKNTIAANS
Component 2
Name:
Trypsin-2
Synonyms:
Anionic trypsinogen | PRSS2 | Serine protease 2 | TRY2 | TRY2_HUMAN | TRYP2 | Thrombin & trypsin | Trypsin | Trypsin II | Trypsin-2
Type:
PROTEIN
Mol. Mass.:
26479.43
Organism:
Homo sapiens (Human)
Description:
ChEMBL_41517
Residue:
247
Sequence:
MNLLLILTFVAAAVAAPFDDDDKIVGGYICEENSVPYQVSLNSGYHFCGGSLISEQWVVSAGHCYKSRIQVRLGEHNIEVLEGNEQFINAAKIIRHPKYNSRTLDNDILLIKLSSPAVINSRVSAISLPTAPPAAGTESLISGWGNTLSSGADYPDELQCLDAPVLSQAECEASYPGKITNNMFCVGFLEGGKDSCQGDSGGPVVSNGELQGIVSWGYGCAQKNRPGVYTKVYNYVDWIKDTIAANS
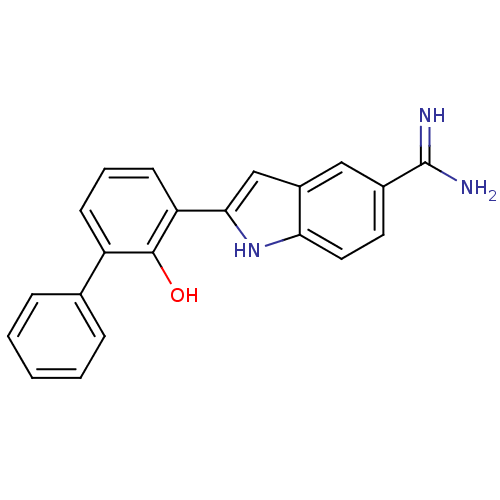
Inhibitor
Name:
BDBM50102780
Synonyms:
2-(2-Hydroxy-3-phenyl-phenyl)-1H-indole-5-carboxamidine | 2-(2-Hydroxy-biphenyl-3-yl)-1H-indole-5-carboxamidine | 3-{5-[AMINO(IMINIO)METHYL]-1H-INDOL-2-YL}-1,1'-BIPHENYL-2-OLATE | CHEMBL96433
Type:
Small organic molecule
Emp. Form.:
C21H17N3O
Mol. Mass.:
327.3792
SMILES:
NC(=N)c1ccc2[nH]c(cc2c1)-c1cccc(-c2ccccc2)c1O