Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Glutamate receptor ionotropic, NMDA 1
Ligand
BDBM50062599
Substrate
n/a
Meas. Tech.
ChEMBL_489454 (CHEMBL982936)
IC50
9520±n/a nM
Citation
 Rosini, M; Simoni, E; Bartolini, M; Cavalli, A; Ceccarini, L; Pascu, N; McClymont, DW; Tarozzi, A; Bolognesi, ML; Minarini, A; Tumiatti, V; Andrisano, V; Mellor, IR; Melchiorre, C Inhibition of acetylcholinesterase, beta-amyloid aggregation, and NMDA receptors in Alzheimer's disease: a promising direction for the multi-target-directed ligands gold rush. J Med Chem 51:4381-4 (2008) [PubMed] Article
Rosini, M; Simoni, E; Bartolini, M; Cavalli, A; Ceccarini, L; Pascu, N; McClymont, DW; Tarozzi, A; Bolognesi, ML; Minarini, A; Tumiatti, V; Andrisano, V; Mellor, IR; Melchiorre, C Inhibition of acetylcholinesterase, beta-amyloid aggregation, and NMDA receptors in Alzheimer's disease: a promising direction for the multi-target-directed ligands gold rush. J Med Chem 51:4381-4 (2008) [PubMed] Article More Info.:
Target
Name:
Glutamate receptor ionotropic, NMDA 1
Synonyms:
GRIN1 | Glutamate (NMDA) receptor subunit zeta 1 | Glutamate [NMDA] receptor subunit zeta-1 | Ionotropic glutamate receptor NMDA 1/2D | N-methyl-D-aspartate receptor subunit NR1 | NMDAR1 | NMDZ1_HUMAN | phencyclidine
Type:
Enzyme Catalytic Domain
Mol. Mass.:
105397.81
Organism:
Homo sapiens (Human)
Description:
Q05586
Residue:
938
Sequence:
MSTMRLLTLALLFSCSVARAACDPKIVNIGAVLSTRKHEQMFREAVNQANKRHGSWKIQLNATSVTHKPNAIQMALSVCEDLISSQVYAILVSHPPTPNDHFTPTPVSYTAGFYRIPVLGLTTRMSIYSDKSIHLSFLRTVPPYSHQSSVWFEMMRVYSWNHIILLVSDDHEGRAAQKRLETLLEERESKAEKVLQFDPGTKNVTALLMEAKELEARVIILSASEDDAATVYRAAAMLNMTGSGYVWLVGEREISGNALRYAPDGILGLQLINGKNESAHISDAVGVVAQAVHELLEKENITDPPRGCVGNTNIWKTGPLFKRVLMSSKYADGVTGRVEFNEDGDRKFANYSIMNLQNRKLVQVGIYNGTHVIPNDRKIIWPGGETEKPRGYQMSTRLKIVTIHQEPFVYVKPTLSDGTCKEEFTVNGDPVKKVICTGPNDTSPGSPRHTVPQCCYGFCIDLLIKLARTMNFTYEVHLVADGKFGTQERVNNSNKKEWNGMMGELLSGQADMIVAPLTINNERAQYIEFSKPFKYQGLTILVKKEIPRSTLDSFMQPFQSTLWLLVGLSVHVVAVMLYLLDRFSPFGRFKVNSEEEEEDALTLSSAMWFSWGVLLNSGIGEGAPRSFSARILGMVWAGFAMIIVASYTANLAAFLVLDRPEERITGINDPRLRNPSDKFIYATVKQSSVDIYFRRQVELSTMYRHMEKHNYESAAEAIQAVRDNKLHAFIWDSAVLEFEASQKCDLVTTGELFFRSGFGIGMRKDSPWKQNVSLSILKSHENGFMEDLDKTWVRYQECDSRSNAPATLTFENMAGVFMLVAGGIVAGIFLIFIEIAYKRHKDARRKQMQLAFAAVNVWRKNLQDRKSGRAEPDPKKKATFRAITSTLASSFKRRRSSKDTSTGGGRGALQNQKDTVLPRRAIEREEGQLQLCSRHRES
Inhibitor
Name:
BDBM50062599
Synonyms:
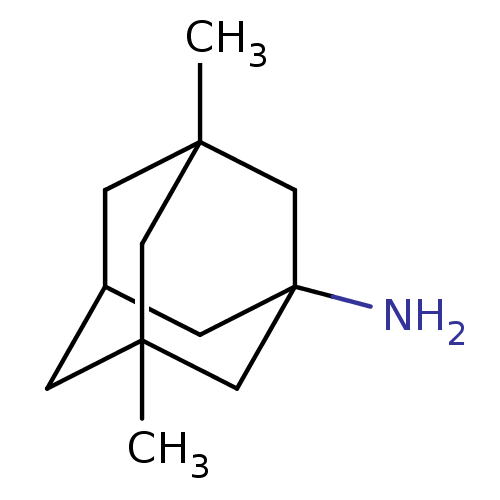
3,5-Dimethyl-adamantan-1-ylamine | CHEMBL807 | EN300-123026 | MEMANTINE | Namenda | US10214478, Compound memantine
Type:
Small organic molecule
Emp. Form.:
C12H21N
Mol. Mass.:
179.3018
SMILES:
CC12CC3CC(C)(C1)CC(N)(C3)C2 |TLB:7:1:4.5.8:11,10:9:4:2.7.1,0:1:4:8.9.11,THB:7:5:11:2.1.12,12:1:4:8.9.11,12:9:4:2.7.1,6:5:11:2.1.12|