Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
cGMP-specific 3',5'-cyclic phosphodiesterase
Ligand
BDBM15296
Substrate
n/a
Meas. Tech.
ChEMBL_155357 (CHEMBL762502)
IC50
>100000±n/a nM
Citation
 Watanabe, N; Adachi, H; Takase, Y; Ozaki, H; Matsukura, M; Miyazaki, K; Ishibashi, K; Ishihara, H; Kodama, K; Nishino, M; Kakiki, M; Kabasawa, Y 4-(3-Chloro-4-methoxybenzyl)aminophthalazines: synthesis and inhibitory activity toward phosphodiesterase 5. J Med Chem 43:2523-9 (2000) [PubMed] Article
Watanabe, N; Adachi, H; Takase, Y; Ozaki, H; Matsukura, M; Miyazaki, K; Ishibashi, K; Ishihara, H; Kodama, K; Nishino, M; Kakiki, M; Kabasawa, Y 4-(3-Chloro-4-methoxybenzyl)aminophthalazines: synthesis and inhibitory activity toward phosphodiesterase 5. J Med Chem 43:2523-9 (2000) [PubMed] Article More Info.:
Target
Name:
cGMP-specific 3',5'-cyclic phosphodiesterase
Synonyms:
3',5'-cyclic phosphodiesterase | CGB-PDE | PDE5 | PDE5A | PDE5A_HUMAN | Phosphodiesterase 2 and 5 (PDE2 and PDE5) | Phosphodiesterase 5 (PDE5) | Phosphodiesterase 5A | Phosphodiesterase 5A (PDE5A) | cGMP-binding cGMP-specific phosphodiesterase | cGMP-specific 3',5'-cyclic phosphodiesterase
Type:
Protein
Mol. Mass.:
99975.83
Organism:
Homo sapiens (Human)
Description:
O76074
Residue:
875
Sequence:
MERAGPSFGQQRQQQQPQQQKQQQRDQDSVEAWLDDHWDFTFSYFVRKATREMVNAWFAERVHTIPVCKEGIRGHTESCSCPLQQSPRADNSAPGTPTRKISASEFDRPLRPIVVKDSEGTVSFLSDSEKKEQMPLTPPRFDHDEGDQCSRLLELVKDISSHLDVTALCHKIFLHIHGLISADRYSLFLVCEDSSNDKFLISRLFDVAEGSTLEEVSNNCIRLEWNKGIVGHVAALGEPLNIKDAYEDPRFNAEVDQITGYKTQSILCMPIKNHREEVVGVAQAINKKSGNGGTFTEKDEKDFAAYLAFCGIVLHNAQLYETSLLENKRNQVLLDLASLIFEEQQSLEVILKKIAATIISFMQVQKCTIFIVDEDCSDSFSSVFHMECEELEKSSDTLTREHDANKINYMYAQYVKNTMEPLNIPDVSKDKRFPWTTENTGNVNQQCIRSLLCTPIKNGKKNKVIGVCQLVNKMEENTGKVKPFNRNDEQFLEAFVIFCGLGIQNTQMYEAVERAMAKQMVTLEVLSYHASAAEEETRELQSLAAAVVPSAQTLKITDFSFSDFELSDLETALCTIRMFTDLNLVQNFQMKHEVLCRWILSVKKNYRKNVAYHNWRHAFNTAQCMFAALKAGKIQNKLTDLEILALLIAALSHDLDHRGVNNSYIQRSEHPLAQLYCHSIMEHHHFDQCLMILNSPGNQILSGLSIEEYKTTLKIIKQAILATDLALYIKRRGEFFELIRKNQFNLEDPHQKELFLAMLMTACDLSAITKPWPIQQRIAELVATEFFDQGDRERKELNIEPTDLMNREKKNKIPSMQVGFIDAICLQLYEALTHVSEDCFPLLDGCRKNRQKWQALAEQQEKMLINGESGQAKRN
Inhibitor
Name:
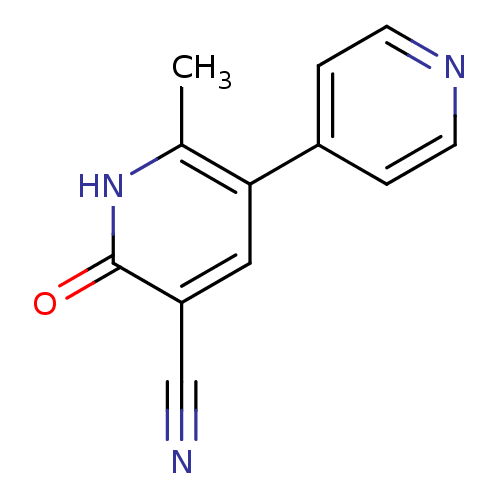
BDBM15296
Synonyms:
6-methyl-2-oxo-5-(pyridin-4-yl)-1,2-dihydropyridine-3-carbonitrile | 6-methyl-2-oxo-5-pyridin-4-yl-1H-pyridine-3-carbonitrile | CHEMBL189 | Milrinone | Primacor | US9242982, Milrinone | US9249139, Milrinone
Type:
Small organic molecule
Emp. Form.:
C12H9N3O
Mol. Mass.:
211.2194
SMILES:
Cc1[nH]c(=O)c(cc1-c1ccncc1)C#N