Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Aurora kinase A
Ligand
BDBM13534
Substrate
n/a
Meas. Tech.
ChEMBL_966729 (CHEMBL2400289)
IC50
20±n/a nM
Citation
 Shiao, HY; Coumar, MS; Chang, CW; Ke, YY; Chi, YH; Chu, CY; Sun, HY; Chen, CH; Lin, WH; Fung, KS; Kuo, PC; Huang, CT; Chang, KY; Lu, CT; Hsu, JT; Chen, CT; Jiaang, WT; Chao, YS; Hsieh, HP Optimization of ligand and lipophilic efficiency to identify an in vivo active furano-pyrimidine Aurora kinase inhibitor. J Med Chem 56:5247-60 (2014) [PubMed] Article
Shiao, HY; Coumar, MS; Chang, CW; Ke, YY; Chi, YH; Chu, CY; Sun, HY; Chen, CH; Lin, WH; Fung, KS; Kuo, PC; Huang, CT; Chang, KY; Lu, CT; Hsu, JT; Chen, CT; Jiaang, WT; Chao, YS; Hsieh, HP Optimization of ligand and lipophilic efficiency to identify an in vivo active furano-pyrimidine Aurora kinase inhibitor. J Med Chem 56:5247-60 (2014) [PubMed] Article More Info.:
Target
Name:
Aurora kinase A
Synonyms:
AIK | AIRK1 | ARK1 | AURA | AURKA | AURKA_HUMAN | AYK1 | Aurora 2 | Aurora kinase A (AURA) | Aurora kinase A (AURKA) | Aurora kinase A (Aurora A) | Aurora kinase A (Aurora-2) | Aurora-related kinase 1 | Aurora/IPL1-related kinase 1 | BTAK | Breast tumor-amplified kinase | Breast-tumor-amplified kinase | IAK1 | STK15 | STK6 | Serine/threonine kinase 15 | Serine/threonine-protein kinase 6 | Serine/threonine-protein kinase aurora A | aurora-2 | hARK1
Type:
Serine/threonine-protein kinase
Mol. Mass.:
45830.98
Organism:
Homo sapiens (Human)
Description:
O14965
Residue:
403
Sequence:
MDRSKENCISGPVKATAPVGGPKRVLVTQQFPCQNPLPVNSGQAQRVLCPSNSSQRVPLQAQKLVSSHKPVQNQKQKQLQATSVPHPVSRPLNNTQKSKQPLPSAPENNPEEELASKQKNEESKKRQWALEDFEIGRPLGKGKFGNVYLAREKQSKFILALKVLFKAQLEKAGVEHQLRREVEIQSHLRHPNILRLYGYFHDATRVYLILEYAPLGTVYRELQKLSKFDEQRTATYITELANALSYCHSKRVIHRDIKPENLLLGSAGELKIADFGWSVHAPSSRRTTLCGTLDYLPPEMIEGRMHDEKVDLWSLGVLCYEFLVGKPPFEANTYQETYKRISRVEFTFPDFVTEGARDLISRLLKHNPSQRPMLREVLEHPWITANSSKPSNCQNKESASKQS
Inhibitor
Name:
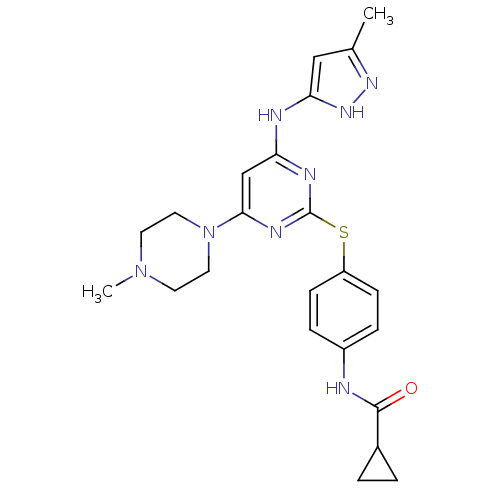
BDBM13534
Synonyms:
CHEMBL572878 | N-[4-({4-[(3-methyl-1H-pyrazol-5-yl)amino]-6-(4-methylpiperazin-1-yl)pyrimidin-2-yl}sulfanyl)phenyl]cyclopropanecarboxamide | N-[4-[[4-(4-methylpiperazino)-6-[(5-methyl-1H-pyrazol-3-yl)amino]pyrimidin-2-yl]thio]phenyl]cyclopropanecarboxamide | VX-680 | VX680 | cyclopropane carboxylic acid {4-[4-(4-methyl-piperazin-1-yl)-6-(5-methyl-2H-pyrazol-3-ylamino)-pyrimidin-2ylsulphanyl]-phenyl}-amide
Type:
Small organic molecule
Emp. Form.:
C23H28N8OS
Mol. Mass.:
464.586
SMILES:
CN1CCN(CC1)c1cc(Nc2cc(C)n[nH]2)nc(Sc2ccc(NC(=O)C3CC3)cc2)n1