Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Phospholipase D2
Ligand
BDBM87146
Substrate
n/a
Meas. Tech.
Cellular PLD2 concentration response
IC50
165±n/a nM
Citation
More Info.:
Target
Name:
Phospholipase D2
Synonyms:
PLD2 | PLD2_HUMAN | Phospholipase D2 | Phospholipase D2 (PLD2) | phospholipase D2 isoform PLD2A
Type:
Protein
Mol. Mass.:
106002.47
Organism:
Homo sapiens (Human)
Description:
n/a
Residue:
933
Sequence:
MTATPESLFPTGDELDSSQLQMESDEVDTLKEGEDPADRMHPFLAIYELQSLKVHPLVFAPGVPVTAQVVGTERYTSGSKVGTCTLYSVRLTHGDFSWTTKKKYRHFQELHRDLLRHKVLMSLLPLARFAVAYSPARDAGNREMPSLPRAGPEGSTRHAASKQKYLENYLNRLLTMSFYRNYHAMTEFLEVSQLSFIPDLGRKGLEGMIRKRSGGHRVPGLTCCGRDQVCYRWSKRWLVVKDSFLLYMCLETGAISFVQLFDPGFEVQVGKRSTEARHGVRIDTSHRSLILKCSSYRQARWWAQEITELAQGPGRDFLQLHRHDSYAPPRPGTLARWFVNGAGYFAAVADAILRAQEEIFITDWWLSPEVYLKRPAHSDDWRLDIMLKRKAEEGVRVSILLFKEVELALGINSGYSKRALMLLHPNIKVMRHPDQVTLWAHHEKLLVVDQVVAFLGGLDLAYGRWDDLHYRLTDLGDSSESAASQPPTPRPDSPATPDLSHNQFFWLGKDYSNLITKDWVQLDRPFEDFIDRETTPRMPWRDVGVVVHGLPARDLARHFIQRWNFTKTTKAKYKTPTYPYLLPKSTSTANQLPFTLPGGQCTTVQVLRSVDRWSAGTLENSILNAYLHTIRESQHFLYIENQFFISCSDGRTVLNKVGDEIVDRILKAHKQGWCYRVYVLLPLLPGFEGDISTGGGNSIQAILHFTYRTLCRGEYSILHRLKAAMGTAWRDYISICGLRTHGELGGHPVSELIYIHSKVLIADDRTVIIGSANINDRSLLGKRDSELAVLIEDTETEPSLMNGAEYQAGRFALSLRKHCFGVILGANTRPDLDLRDPICDDFFQLWQDMAESNANIYEQIFRCLPSNATRSLRTLREYVAVEPLATVSPPLARSELTQVQGHLVHFPLKFLEDESLLPPLGSKEGMIPLEVWT
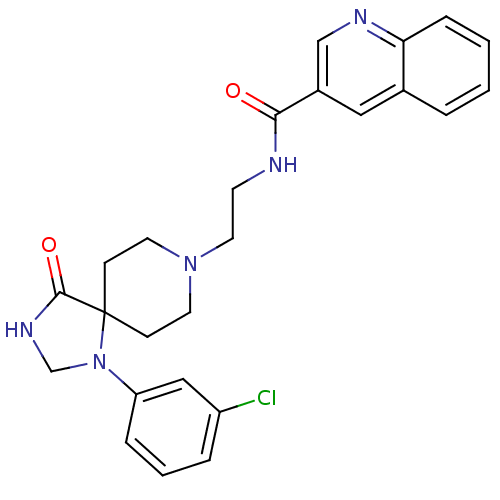
Inhibitor
Name:
BDBM87146
Synonyms:
CHEMBL1254822 | N-[2-[1-(3-chlorophenyl)-4-keto-1,3,8-triazaspiro[4.5]decan-8-yl]ethyl]quinoline-3-carboxamide;2,2,2-trifluoroacetic acid | N-[2-[1-(3-chlorophenyl)-4-oxidanylidene-1,3,8-triazaspiro[4.5]decan-8-yl]ethyl]quinoline-3-carboxamide;2,2,2-tris(fluoranyl)ethanoic acid | N-[2-[1-(3-chlorophenyl)-4-oxo-1,3,8-triazaspiro[4.5]decan-8-yl]ethyl]-3-quinolinecarboxamide;2,2,2-trifluoroacetic acid | N-[2-[1-(3-chlorophenyl)-4-oxo-1,3,8-triazaspiro[4.5]decan-8-yl]ethyl]quinoline-3-carboxamide;2,2,2-trifluoroacetic acid | VU0405263-1 | cid_53469923
Type:
Small organic molecule
Emp. Form.:
C25H26ClN5O2
Mol. Mass.:
463.959
SMILES:
Clc1cccc(c1)N1CNC(=O)C11CCN(CCNC(=O)c2cnc3ccccc3c2)CC1