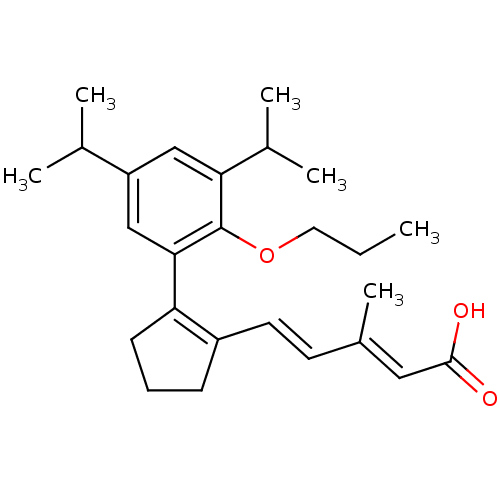
BDBM50133126 5-[2-(3,5-Diisopropyl-2-propoxy-phenyl)-cyclopent-1-enyl]-3-methyl-penta-2,4-dienoic acid::CHEMBL133176
SMILES CCCOc1c(cc(cc1C1=C(CCC1)\C=C\C(\C)=C\C(O)=O)C(C)C)C(C)C
InChI Key InChIKey=AEXZEXFZLVOUCR-OTIKAVQOSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 9 hits for monomerid = 50133126
Found 9 hits for monomerid = 50133126
Affinity DataKi: 1.90nMAssay Description:Binding affinity for Retinoic acid receptor RXR-alpha was determined by competing with 3[H]-9-cis-RAMore data for this Ligand-Target Pair
Affinity DataKi: 4.90nMAssay Description:Binding affinity for Retinoic acid receptor RXR-gamma was determined by competing with 3[H]-9-cis-RAMore data for this Ligand-Target Pair
Affinity DataKi: 6nMAssay Description:Binding affinity for Retinoic acid receptor RXR-beta was determined by competing with 3[H]-9-cis-RAMore data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor alpha(Rattus norvegicus)
Ligand Pharmaceuticals
Curated by ChEMBL
Ligand Pharmaceuticals
Curated by ChEMBL
Affinity DataKi: >1.00E+3nMAssay Description:Binding affinity for peroxisome Peroxisome proliferator activated receptor alpha was determinedMore data for this Ligand-Target Pair
Affinity DataKi: >1.00E+3nMAssay Description:Binding affinity for Retinoic acid receptor gamma was determined by competing with 3[H]-ATRAMore data for this Ligand-Target Pair
TargetRetinoic acid receptor, alpha, isoform CRA_b(Rattus norvegicus)
Ligand Pharmaceuticals
Curated by ChEMBL
Ligand Pharmaceuticals
Curated by ChEMBL
Affinity DataKi: 1.40E+3nMAssay Description:Binding affinity for Retinoic acid receptor alpha was determined by competing with 3[H]-ATRAMore data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor gamma(Rattus norvegicus)
Ligand Pharmaceuticals
Curated by ChEMBL
Ligand Pharmaceuticals
Curated by ChEMBL
Affinity DataKi: 3.22E+3nMAssay Description:Binding affinity for peroxisome proliferator activated receptor gammaMore data for this Ligand-Target Pair
Affinity DataKi: 5.62E+3nMAssay Description:Binding affinity for Retinoic acid receptor beta was determined by competing with 3[H]-ATRAMore data for this Ligand-Target Pair
Affinity DataIC50: 4.5nMAssay Description:Molar concentration required to inhibit 50% of the activating delayed-rectifier K+ current in isolated guinea pig ventricularmyocytesMore data for this Ligand-Target Pair