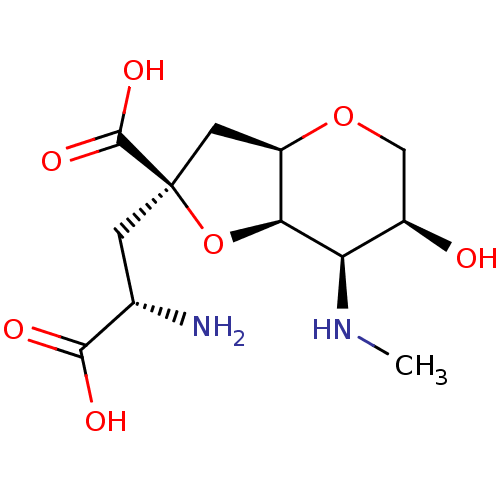
BDBM85740 Dysiherbaine
SMILES CN[C@H]1[C@H]2O[C@](C[C@H](N)C(O)=O)(C[C@H]2OC[C@H]1O)C(O)=O
InChI Key InChIKey=YUSZFKPLFIQTGF-FDNSHYBFSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 5 hits for monomerid = 85740
Found 5 hits for monomerid = 85740
TargetGlutamate receptor ionotropic, kainate 1(Homo sapiens (Human))
Yokohama City University
Curated by ChEMBL
Yokohama City University
Curated by ChEMBL
Affinity DataKi: 0.740nMAssay Description:Displacement of [3H]kianic acid from recombinant GluK1 kainate receptor (unknown origin) expressed in HEK293 cell membranes measured after 1 hrMore data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, kainate 2(Homo sapiens (Human))
Yokohama City University
Curated by ChEMBL
Yokohama City University
Curated by ChEMBL
Affinity DataKi: 1.20nMAssay Description:Displacement of [3H]kianic acid from recombinant GluK2 kainate receptor (unknown origin) expressed in HEK293 cell membranes measured after 1 hrMore data for this Ligand-Target Pair
 3D Structure (crystal)
3D Structure (crystal)