Discovery of Tyrosine Kinase 2 (TYK2) Inhibitor (PF-06826647) for the Treatment of Autoimmune Diseases.
Gerstenberger, B.S., Ambler, C., Arnold, E.P., Banker, M.E., Brown, M.F., Clark, J.D., Dermenci, A., Dowty, M.E., Fensome, A., Fish, S., Hayward, M.M., Hegen, M., Hollingshead, B.D., Knafels, J.D., Lin, D.W., Lin, T.H., Owen, D.R., Saiah, E., Sharma, R., Vajdos, F.F., Xing, L., Yang, X., Yang, X., Wright, S.W.(2020) J Med Chem 63: 13561-13577
- PubMed: 32787094
- DOI: https://doi.org/10.1021/acs.jmedchem.0c00948
- Primary Citation of Related Structures:
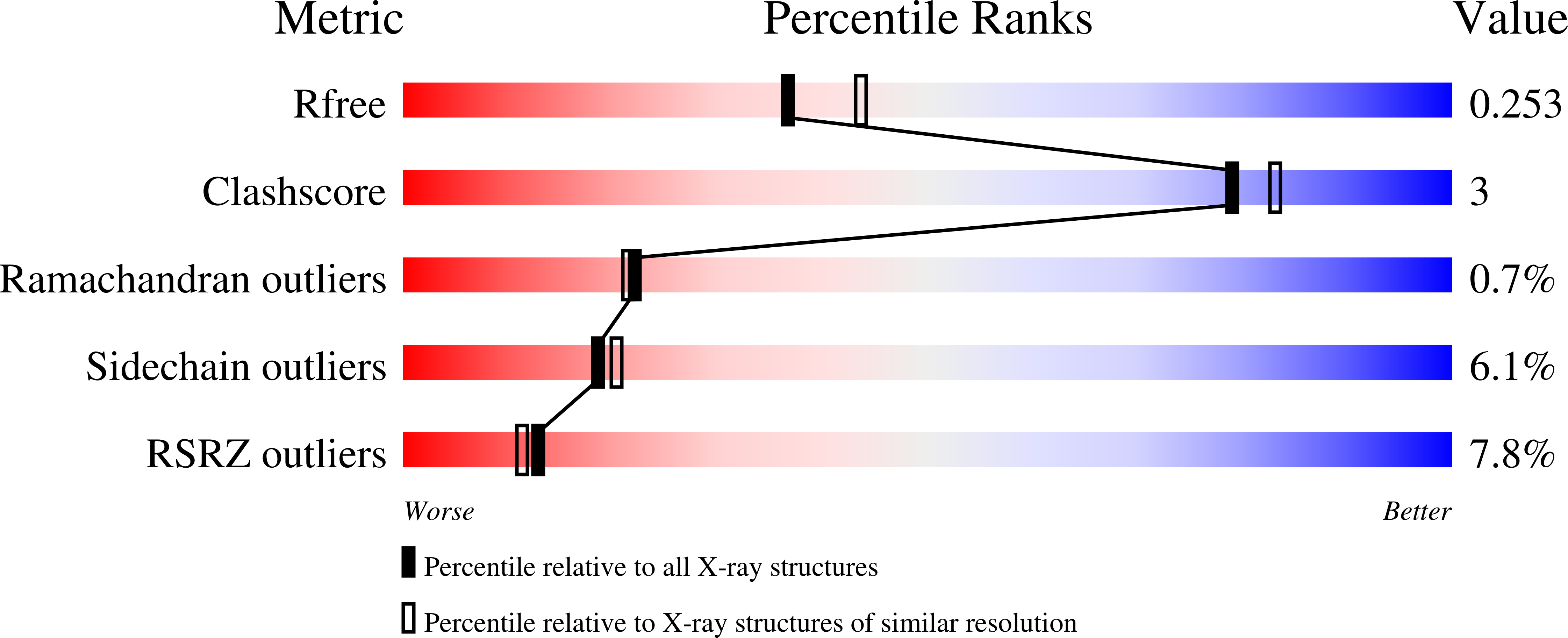
6X8E, 6X8F, 6X8G - PubMed Abstract:
Tyrosine kinase 2 (TYK2) is a member of the JAK kinase family that regulates signal transduction downstream of receptors for the IL-23/IL-12 pathways and type I interferon family, where it pairs with JAK2 or JAK1, respectively. On the basis of human genetic and emerging clinical data, a selective TYK2 inhibitor provides an opportunity to treat autoimmune diseases delivering a potentially differentiated clinical profile compared to currently approved JAK inhibitors. The discovery of an ATP-competitive pyrazolopyrazinyl series of TYK2 inhibitors was accomplished through computational and structurally enabled design starting from a known kinase hinge binding motif. With understanding of PK/PD relationships, a target profile balancing TYK2 potency and selectivity over off-target JAK2 was established. Lead optimization involved modulating potency, selectivity, and ADME properties which led to the identification of the clinical candidate PF-06826647 ( 22 ).
Organizational Affiliation:
Pfizer Inc., Cambridge, Massachusetts 02139, United States.