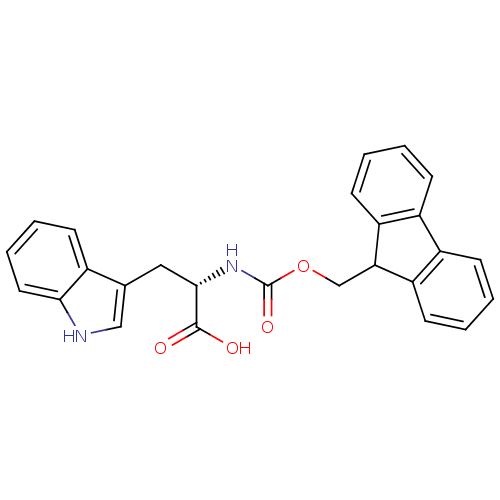
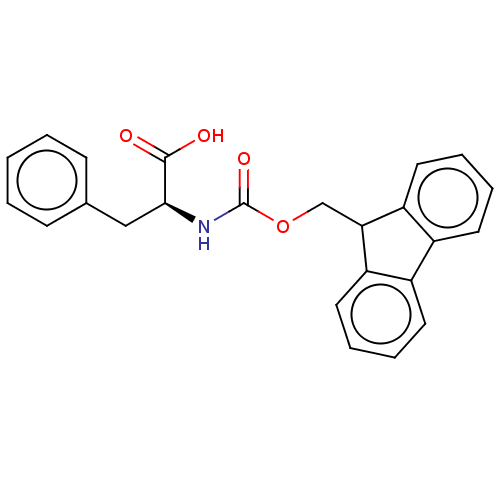
Affinity DataIC50: 2.66E+3nMAssay Description:Inhibition of rhodamine-WEYIPNV binding to Escherichia coli SurA by fluorescence anisotropyMore data for this Ligand-Target Pair
Affinity DataIC50: 2.33E+4nMAssay Description:Inhibition of rhodamine-WEYIPNV binding to Escherichia coli SurA by fluorescence anisotropyMore data for this Ligand-Target Pair
Affinity DataIC50: 3.74E+4nMAssay Description:Inhibition of rhodamine-WEYIPNV binding to Escherichia coli SurA in presence of 0.01% Triton-X-100 by fluorescence anisotropyMore data for this Ligand-Target Pair
Affinity DataIC50: 5.70E+4nMAssay Description:Inhibition of rhodamine-WEYIPNV binding to Escherichia coli SurA by fluorescence anisotropyMore data for this Ligand-Target Pair
Affinity DataIC50: 1.52E+5nMAssay Description:Inhibition of rhodamine-WEYIPNV binding to Escherichia coli SurA by fluorescence anisotropyMore data for this Ligand-Target Pair