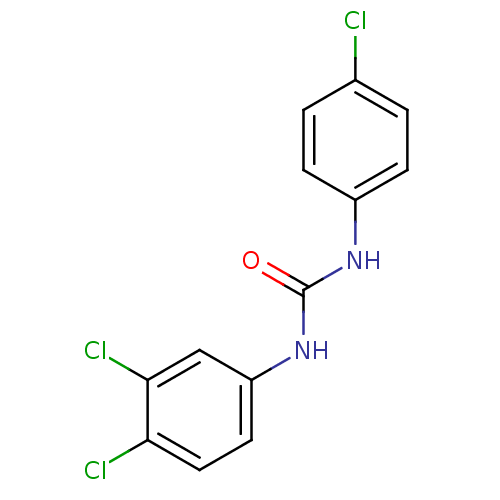
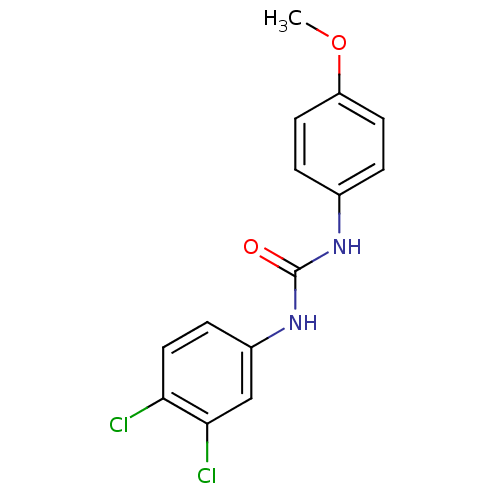
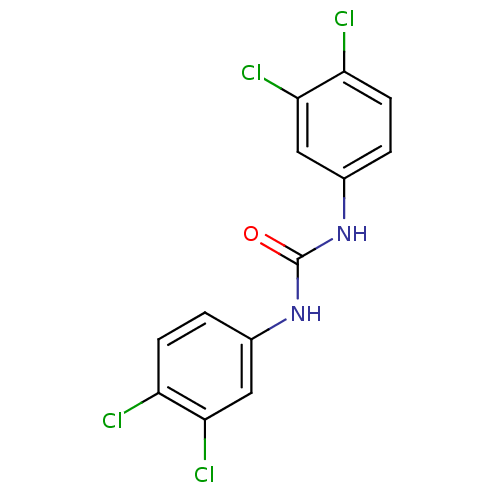
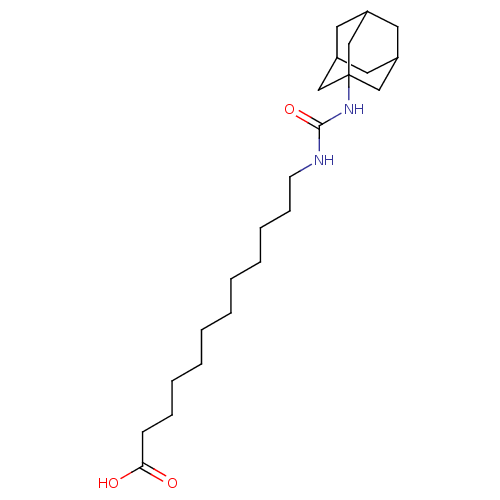
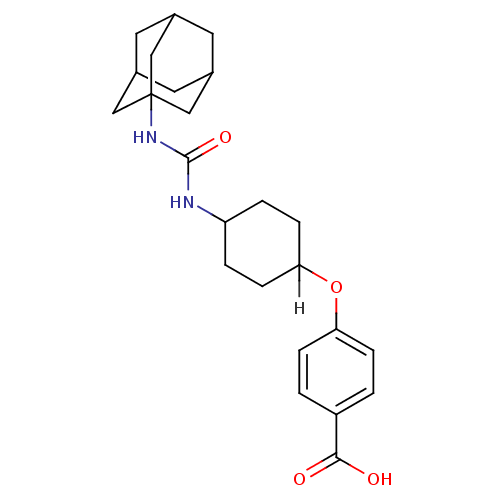
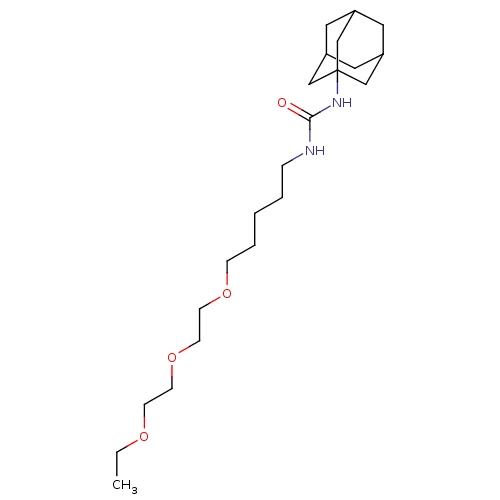
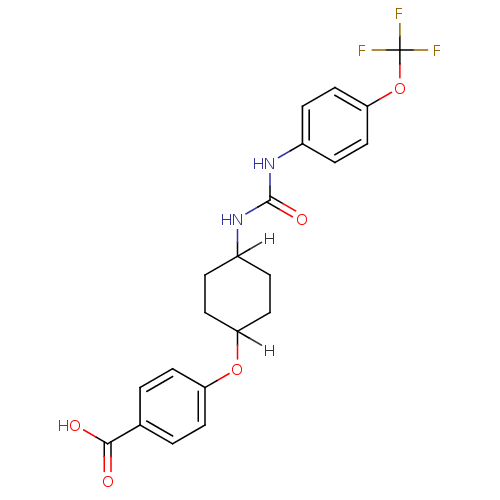
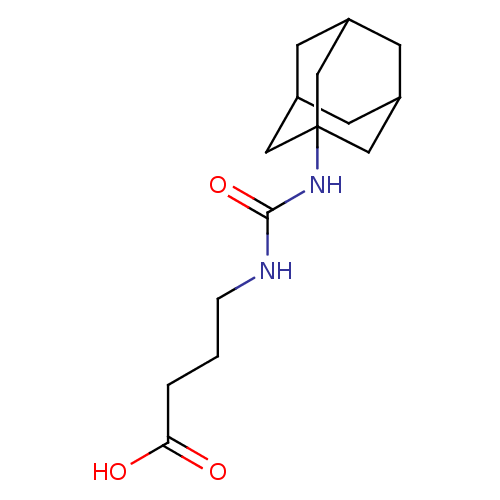
Affinity DataIC50: 19nMpH: 7.0 T: 2°CAssay Description:Enzyme activity was measured by monitoring the appearance of fluorescent product, 6-methoxy-naphthaldehyde (ex@330 nm and em@ 465 nm) on a SpectraMax...More data for this Ligand-Target Pair
Affinity DataIC50: 19nMAssay Description:Inhibition of Mycobacterium tuberculosis recombinant EphB expressed in Escherichia coli BL21 using CMNPC as substrate after 30 mins by fluorescent as...More data for this Ligand-Target Pair
Affinity DataIC50: 30nMpH: 7.0 T: 2°CAssay Description:Enzyme activity was measured by monitoring the appearance of fluorescent product, 6-methoxy-naphthaldehyde (ex@330 nm and em@ 465 nm) on a SpectraMax...More data for this Ligand-Target Pair
Affinity DataIC50: 130nMpH: 7.0 T: 2°CAssay Description:Enzyme activity was measured by monitoring the appearance of fluorescent product, 6-methoxy-naphthaldehyde (ex@330 nm and em@ 465 nm) on a SpectraMax...More data for this Ligand-Target Pair
Affinity DataIC50: 130nMAssay Description:Inhibition of Mycobacterium tuberculosis recombinant EphB expressed in Escherichia coli BL21 using CMNPC as substrate after 30 mins by fluorescent as...More data for this Ligand-Target Pair
Affinity DataIC50: 170nMpH: 7.0 T: 2°CAssay Description:Enzyme activity was measured by monitoring the appearance of fluorescent product, 6-methoxy-naphthaldehyde (ex@330 nm and em@ 465 nm) on a SpectraMax...More data for this Ligand-Target Pair
Affinity DataIC50: 220nMAssay Description:Inhibition of Mycobacterium tuberculosis recombinant EphB expressed in Escherichia coli BL21 using CMNPC as substrate after 30 mins by fluorescent as...More data for this Ligand-Target Pair
Affinity DataIC50: 220nMpH: 7.0 T: 2°CAssay Description:Enzyme activity was measured by monitoring the appearance of fluorescent product, 6-methoxy-naphthaldehyde (ex@330 nm and em@ 465 nm) on a SpectraMax...More data for this Ligand-Target Pair
Affinity DataIC50: 290nMpH: 7.0 T: 2°CAssay Description:Enzyme activity was measured by monitoring the appearance of fluorescent product, 6-methoxy-naphthaldehyde (ex@330 nm and em@ 465 nm) on a SpectraMax...More data for this Ligand-Target Pair
Affinity DataIC50: 460nMpH: 7.0 T: 2°CAssay Description:Enzyme activity was measured by monitoring the appearance of fluorescent product, 6-methoxy-naphthaldehyde (ex@330 nm and em@ 465 nm) on a SpectraMax...More data for this Ligand-Target Pair
Affinity DataIC50: 480nMpH: 7.0 T: 2°CAssay Description:Enzyme activity was measured by monitoring the appearance of fluorescent product, 6-methoxy-naphthaldehyde (ex@330 nm and em@ 465 nm) on a SpectraMax...More data for this Ligand-Target Pair
Affinity DataIC50: 1.96E+3nMpH: 7.0 T: 2°CAssay Description:Enzyme activity was measured by monitoring the appearance of fluorescent product, 6-methoxy-naphthaldehyde (ex@330 nm and em@ 465 nm) on a SpectraMax...More data for this Ligand-Target Pair
Affinity DataIC50: 2.06E+3nMpH: 7.0 T: 2°CAssay Description:Enzyme activity was measured by monitoring the appearance of fluorescent product, 6-methoxy-naphthaldehyde (ex@330 nm and em@ 465 nm) on a SpectraMax...More data for this Ligand-Target Pair
Affinity DataIC50: 3.40E+3nMpH: 7.0 T: 2°CAssay Description:Enzyme activity was measured by monitoring the appearance of fluorescent product, 6-methoxy-naphthaldehyde (ex@330 nm and em@ 465 nm) on a SpectraMax...More data for this Ligand-Target Pair
Affinity DataIC50: 3.53E+3nMpH: 7.0 T: 2°CAssay Description:Enzyme activity was measured by monitoring the appearance of fluorescent product, 6-methoxy-naphthaldehyde (ex@330 nm and em@ 465 nm) on a SpectraMax...More data for this Ligand-Target Pair
Affinity DataIC50: 8.20E+3nMpH: 7.0 T: 2°CAssay Description:Enzyme activity was measured by monitoring the appearance of fluorescent product, 6-methoxy-naphthaldehyde (ex@330 nm and em@ 465 nm) on a SpectraMax...More data for this Ligand-Target Pair
Affinity DataIC50: 1.21E+4nMpH: 7.0 T: 2°CAssay Description:Enzyme activity was measured by monitoring the appearance of fluorescent product, 6-methoxy-naphthaldehyde (ex@330 nm and em@ 465 nm) on a SpectraMax...More data for this Ligand-Target Pair
Affinity DataIC50: 1.67E+4nMpH: 7.0 T: 2°CAssay Description:Enzyme activity was measured by monitoring the appearance of fluorescent product, 6-methoxy-naphthaldehyde (ex@330 nm and em@ 465 nm) on a SpectraMax...More data for this Ligand-Target Pair
Affinity DataIC50: 1.85E+4nMpH: 7.0 T: 2°CAssay Description:Enzyme activity was measured by monitoring the appearance of fluorescent product, 6-methoxy-naphthaldehyde (ex@330 nm and em@ 465 nm) on a SpectraMax...More data for this Ligand-Target Pair
Affinity DataIC50: 2.79E+4nMpH: 7.0 T: 2°CAssay Description:Enzyme activity was measured by monitoring the appearance of fluorescent product, 6-methoxy-naphthaldehyde (ex@330 nm and em@ 465 nm) on a SpectraMax...More data for this Ligand-Target Pair
Affinity DataIC50: 3.16E+4nMpH: 7.0 T: 2°CAssay Description:Enzyme activity was measured by monitoring the appearance of fluorescent product, 6-methoxy-naphthaldehyde (ex@330 nm and em@ 465 nm) on a SpectraMax...More data for this Ligand-Target Pair
Affinity DataIC50: 3.16E+4nMAssay Description:Inhibition of Mycobacterium tuberculosis recombinant EphB expressed in Escherichia coli BL21 using CMNPC as substrate after 30 mins by fluorescent as...More data for this Ligand-Target Pair
Affinity DataIC50: 3.49E+4nMpH: 7.0 T: 2°CAssay Description:Enzyme activity was measured by monitoring the appearance of fluorescent product, 6-methoxy-naphthaldehyde (ex@330 nm and em@ 465 nm) on a SpectraMax...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.0 T: 2°CAssay Description:Enzyme activity was measured by monitoring the appearance of fluorescent product, 6-methoxy-naphthaldehyde (ex@330 nm and em@ 465 nm) on a SpectraMax...More data for this Ligand-Target Pair


 3D Structure (crystal)
3D Structure (crystal)