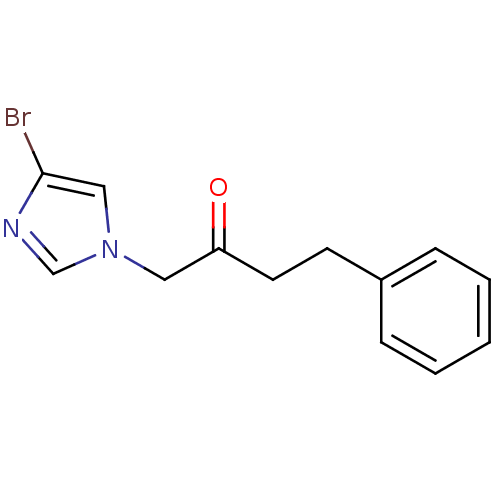
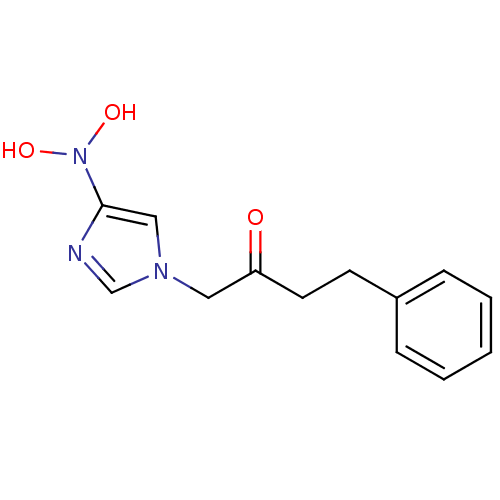
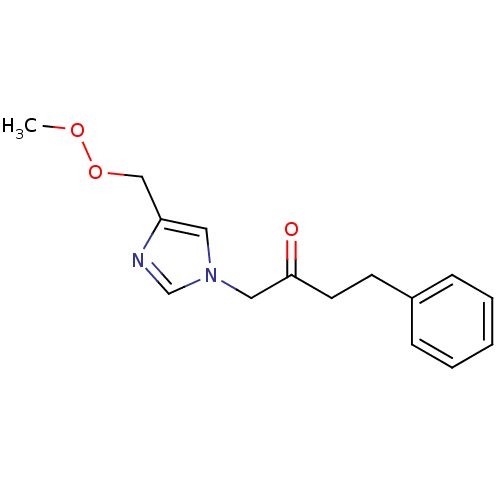
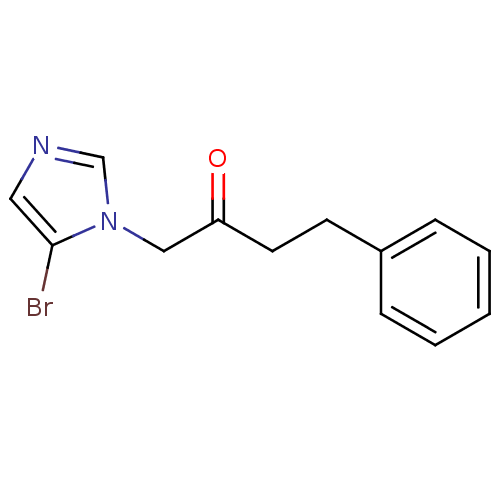
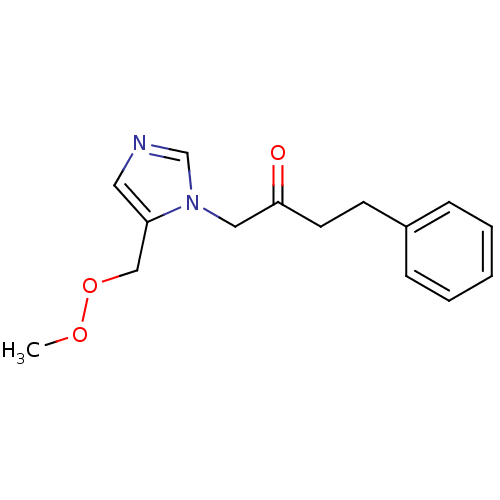
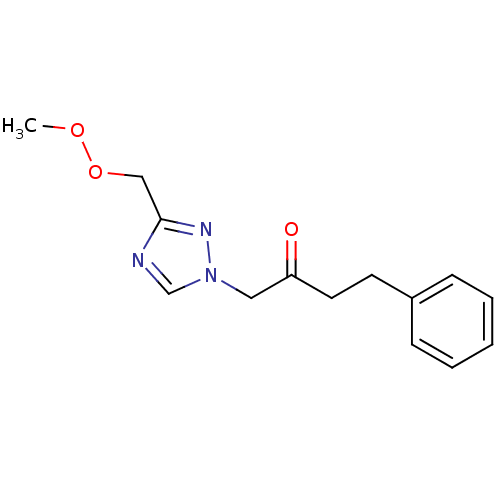
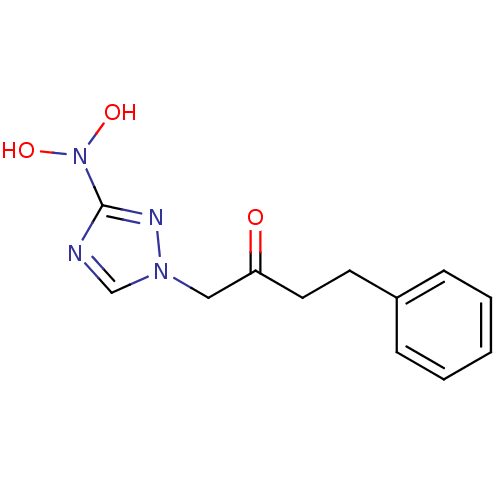
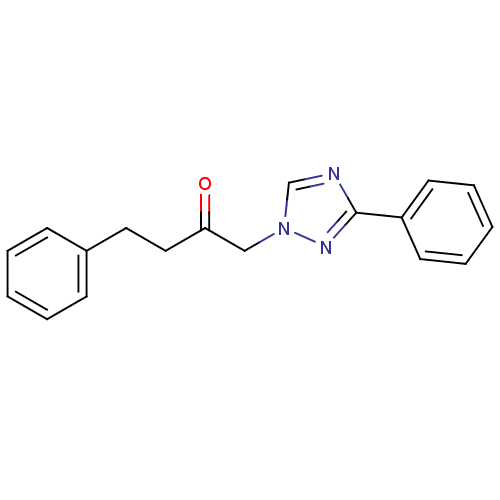
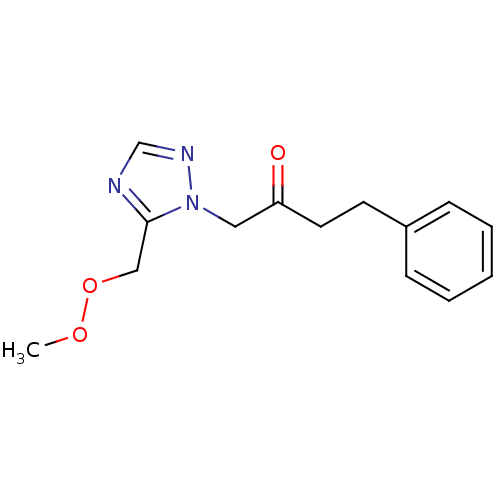
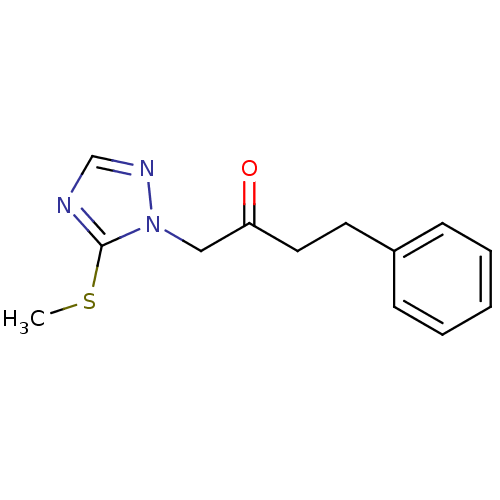
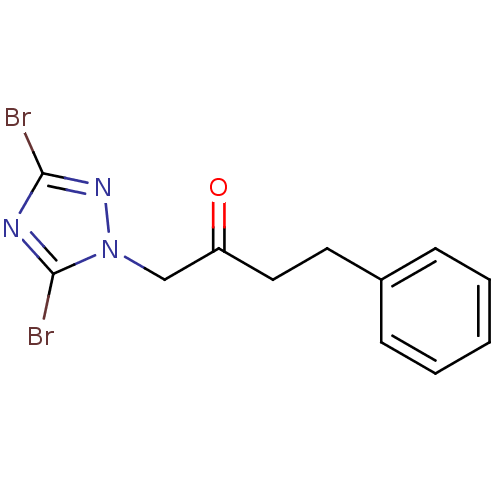
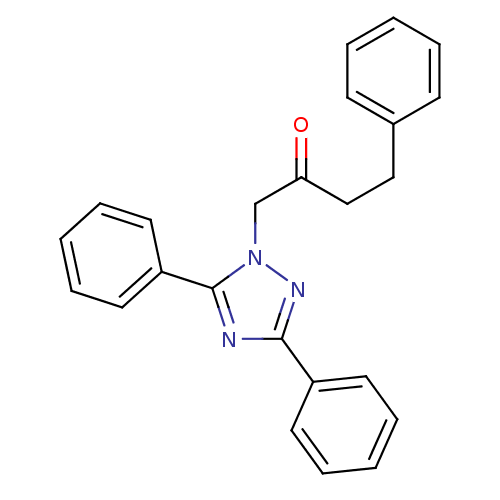
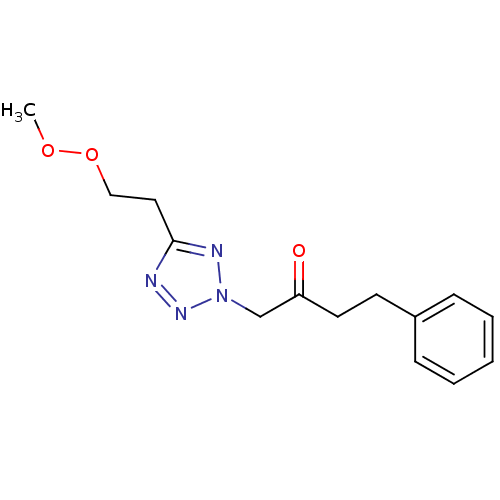
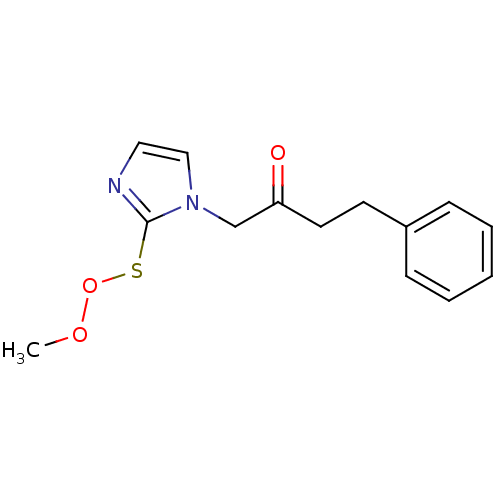
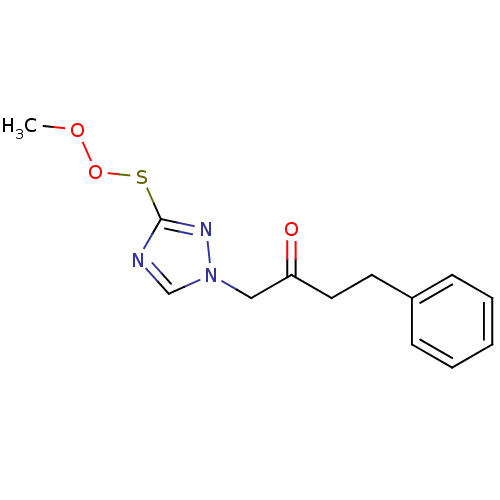
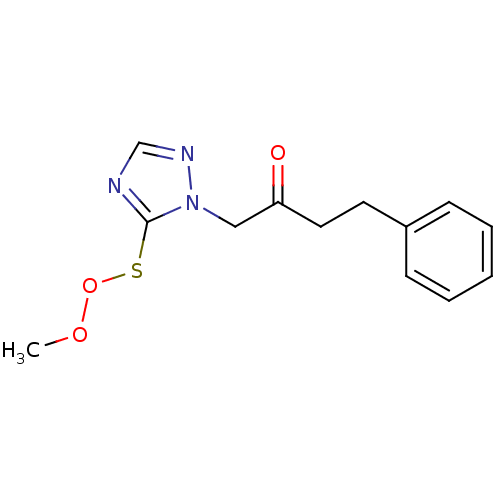
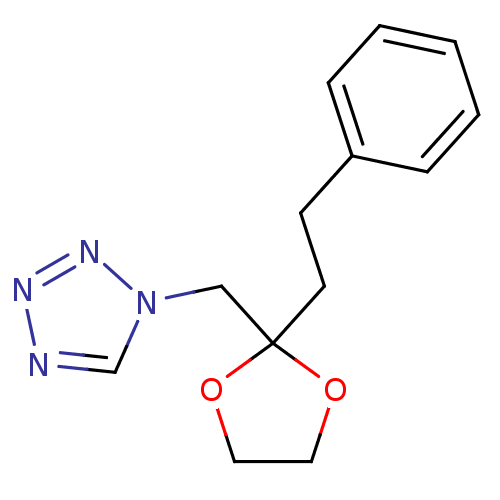
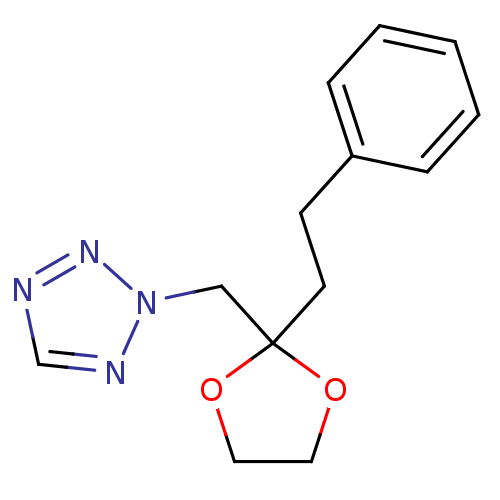
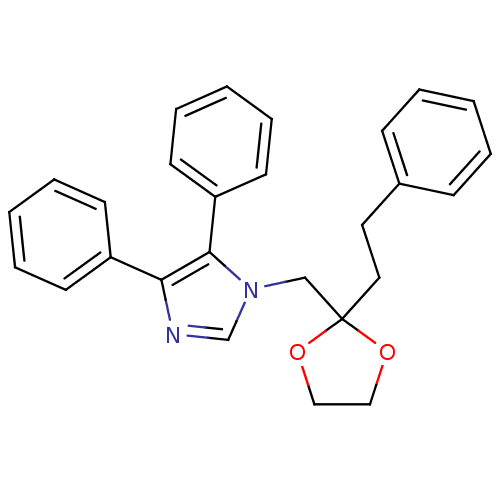
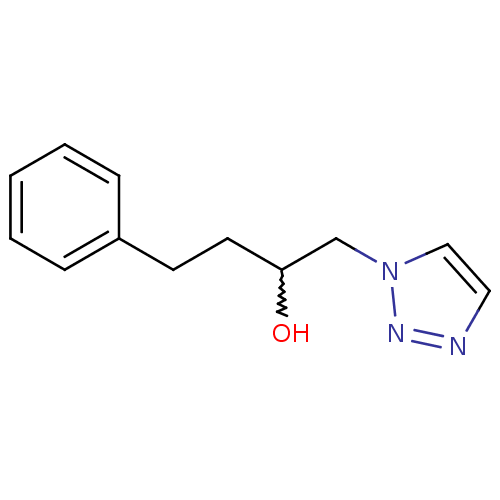
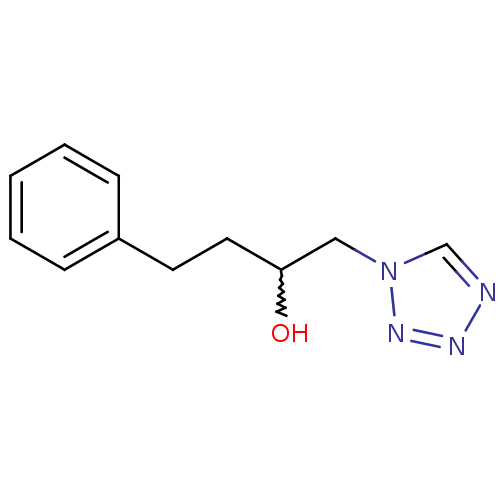
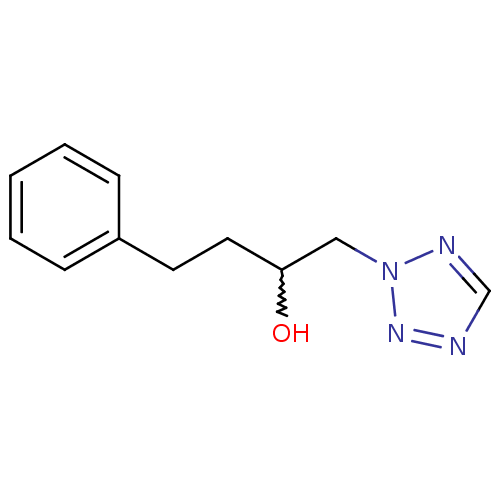
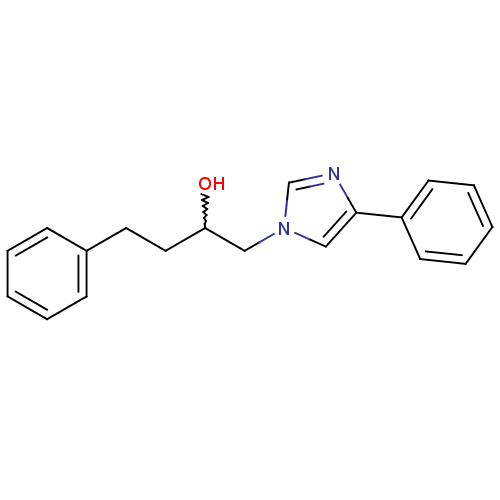
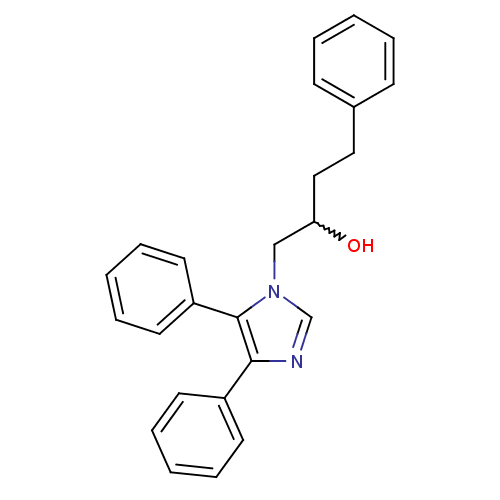
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: 9.00E+4nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: 1.60E+4nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: 5.40E+4nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+5nMpH: 7.4 T: 2°CAssay Description:Heme oxygenase activity in rat spleen and brain microsomal fractions was determined by the quantitation of CO formed from the degradation of methemal...More data for this Ligand-Target Pair
Affinity DataIC50: 3.40E+4nMpH: 7.4Assay Description:HO activity in rat spleen (HO-1) and brain (HO-2) microsomal fractions was determined by the quantitation of CO formed from the degradation of methem...More data for this Ligand-Target Pair
Affinity DataIC50: 8.00E+3nMpH: 7.4Assay Description:HO activity in rat spleen (HO-1) and brain (HO-2) microsomal fractions was determined by the quantitation of CO formed from the degradation of methem...More data for this Ligand-Target Pair
Affinity DataIC50: 4.20E+4nMpH: 7.4Assay Description:HO activity in rat spleen (HO-1) and brain (HO-2) microsomal fractions was determined by the quantitation of CO formed from the degradation of methem...More data for this Ligand-Target Pair
Affinity DataIC50: 6.00E+3nMpH: 7.4Assay Description:HO activity in rat spleen (HO-1) and brain (HO-2) microsomal fractions was determined by the quantitation of CO formed from the degradation of methem...More data for this Ligand-Target Pair
Affinity DataIC50: 6.90E+4nMpH: 7.4Assay Description:HO activity in rat spleen (HO-1) and brain (HO-2) microsomal fractions was determined by the quantitation of CO formed from the degradation of methem...More data for this Ligand-Target Pair
Affinity DataIC50: 7.00E+3nMpH: 7.4Assay Description:HO activity in rat spleen (HO-1) and brain (HO-2) microsomal fractions was determined by the quantitation of CO formed from the degradation of methem...More data for this Ligand-Target Pair
Affinity DataIC50: 2.40E+3nMpH: 7.4Assay Description:HO activity in rat spleen (HO-1) and brain (HO-2) microsomal fractions was determined by the quantitation of CO formed from the degradation of methem...More data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+3nMpH: 7.4Assay Description:HO activity in rat spleen (HO-1) and brain (HO-2) microsomal fractions was determined by the quantitation of CO formed from the degradation of methem...More data for this Ligand-Target Pair
Affinity DataIC50: 1.20E+4nMpH: 7.4Assay Description:HO activity in rat spleen (HO-1) and brain (HO-2) microsomal fractions was determined by the quantitation of CO formed from the degradation of methem...More data for this Ligand-Target Pair
Affinity DataIC50: 5.00E+3nMpH: 7.4Assay Description:HO activity in rat spleen (HO-1) and brain (HO-2) microsomal fractions was determined by the quantitation of CO formed from the degradation of methem...More data for this Ligand-Target Pair
Affinity DataIC50: 1.90E+4nMpH: 7.4Assay Description:HO activity in rat spleen (HO-1) and brain (HO-2) microsomal fractions was determined by the quantitation of CO formed from the degradation of methem...More data for this Ligand-Target Pair